
Updated on March 2, 2023
Rare diseases are defined as diseases that affect a small number of people — fewer than 1 in 2,000 in the European Union and fewer than 200,000 total people (about 1 in 1,500) in the United States. For example, Tay-Sachs disease affects 1 in 300,000 while cystic fibrosis is more common and affects 1 in 10,000. Though individual rare diseases affect very few people, collectively they are common and affect over 300 million people worldwide.
Advances in sequencing technology improve our understanding of rare diseases
With more than 70% of rare diseases being genetic in origin, scientists around the world have deployed genomic technologies to identify their causal mechanisms. Improvement in the technologies for identifying genetic variation have increased scientists’ ability to understand rare diseases. Learn more about the evolution of DNA sequencing tools.
Karyotyping was the first technology to provide a view of the genome, revealing diseases due to chromosomal abnormalities such as Turner syndrome (1 chromosome X instead of 2 in a female). Later, microarray provided a higher-resolution view, identifying large copy number variants, as in DiGeorge syndrome (caused by a deletion of around 2.5 Mb on chromosome 22). Exome, or whole genome sequencing based on short-read sequencing platforms, enabled even more progress by detecting single nucleotide variants (SNVs), insertions and deletions, and some larger variants.
But even whole genome sequencing with short reads finds a genetic cause in less than half of all instances of rare disease — leaving the causes of many rare diseases unknown. This in part is because even whole genome sequencing with short reads does not provide a comprehensive view of variation.
Fortunately, more recent advancements have led to the introduction of long-read sequencing, which has enabled sequencing of the whole human genome — every single base — so that all types of variants can be detected from SNV up to large structural variants (SVs). Ultimately, by detecting more variants, long-read sequencing provides a more complete picture of the genome and any abnormalities that may exist.
In case you missed it: reaching a genomics milestone — the first complete human genome.
What’s the difference between short-read sequencing and long-read sequencing?
Like their names suggest, short-read sequencing looks at DNA in short snippets (100–350 base pairs) while long-read sequencing measures long fragments of DNA (tens of thousands of base pairs). Why does that matter? Well, when trying to characterize a human genome that has two copies (one maternal and one paternal), each 3.2 billion base pairs in length. Having longer snippets of DNA means you:
- Need fewer snippets to make up the length of the whole genome and have no gaps where the sequence is unknown
- Can more easily map how one region of the genome is connected to another region
- Have the ability to phase or determine which copy of a gene, maternal or paternal, a mutation occurs in
As it turns out, the genetic variants underlying many of these diseases are exactly the types that short-read sequencers are least able to detect. From repeat expansions to large deletions or insertions, pathogenic variants are often large and complex structural elements that cannot be spanned by short reads of just a few hundred bases. Representing these variants accurately — and capturing all types of variants — requires much longer sequence reads that cover the entire variant in a single stretch.
HiFi sequencing — the key to seeing all variant types involved in rare disease
Unlike the data produced by short-read sequencing platforms, highly accurate long-read sequencing, known as HiFi sequencing, generates extremely long reads (>25 kb) that span even the largest structural variants. HiFi sequencing provides the most comprehensive view of variation in a genome, identifying the variation found with short reads and detecting the larger and more complex variants that short reads miss.
The long reads and high accuracy (>99.9%) of HiFi sequencing provide very complete genome assemblies, comprehensive variant detection with base-pair resolution, and phasing to represent maternal and paternal haplotypes.
Unlock the secrets of rare disease with HiFi sequencing
HiFi sequencing has already made a substantial difference in rare disease research by identifying variants that were missed by short-read sequencing and other technologies. For more detail, check out these research studies of undiagnosed rare diseases and the types of pathogenic variants underlying them.
Structural variant calling in rare disease studies
One of the earliest examples of how PacBio sequencing technology could play a role in rare disease research came from the Stanford lab of cardiologist Euan Ashley (@euanashley) and a young man who had suffered a series of tumors in his heart and glands. Eight years of genetic analyses had produced no firm answers. Ashley’s team used a novel method of PacBio whole genome sequencing to find a novel structural variant in a gene associated with Carney syndrome, which was later validated as the correct mutation and finding.
More recently, a group at HudsonAlpha found new evidence in the study of a young girl with intellectual disabilities, seizures, and speech delay. With HiFi sequencing, the scientists at HudsonAlpha identified a de novo heterozygous insertion of nearly 7,000 bases in an intron of the CDKL5 gene that they deemed likely pathogenic. Since CDKL5 has been associated with early infantile epileptic encephalopathy 2, a condition characterized by many symptoms experienced by the proband, “we prioritized this event as the most interesting candidate variant,” the authors reported.
Structural variants are generally classified as being >50 bp in length and include insertions, deletions, duplications, copy-number variants, inversions, and translocations. Learn more.
In Japan, researchers deployed HiFi sequencing to find the cause of an undiagnosed syndrome in twin 12-year-old girls. Clinical symptoms matched Dravet syndrome, but no molecular evidence was available to confirm that finding. They sequenced one of the twins and both parents, identifying a novel 12 kb inversion in a region that had previously been associated with the same symptoms affecting the girls.
In one last structural variant example, Kristen Sund (@kristen_sund) from Cincinnati Children’s Hospital identified a 13 Mb complex rearrangement that appears to be responsible for a movement disorder in a 17-year-old with chorea, myoclonus, anxiety, and hypothyroidism. The variant was found in the NKX2-1 gene.
Small variants in challenging regions of the genome
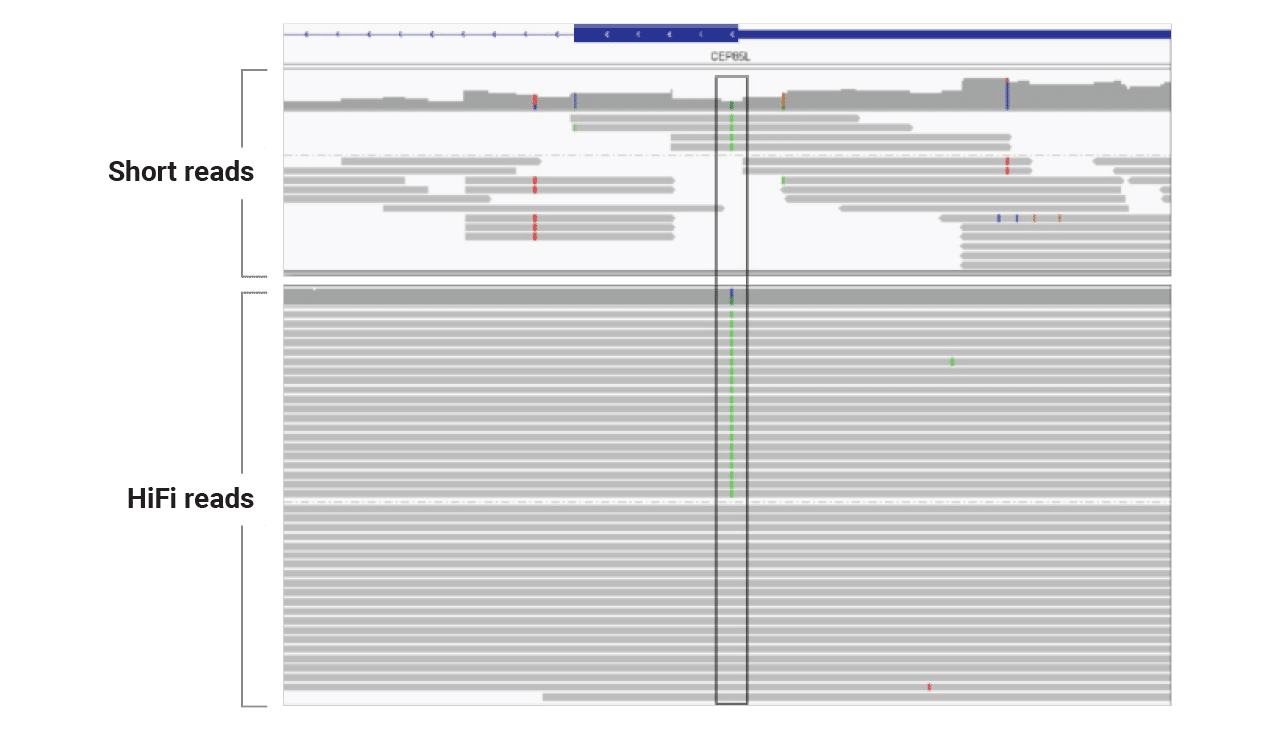
For an individual with lissencephaly (lack of folds in brain), developmental delay, and seizures, scientists at Children’s Mercy Kansas City used HiFi sequencing to reveal a pathogenic variant in a region that proved difficult for short reads to represent accurately. HiFi sequencing provided even coverage — unlike the coverage dropout seen with short-read data for the same region — which spotted the key variant.
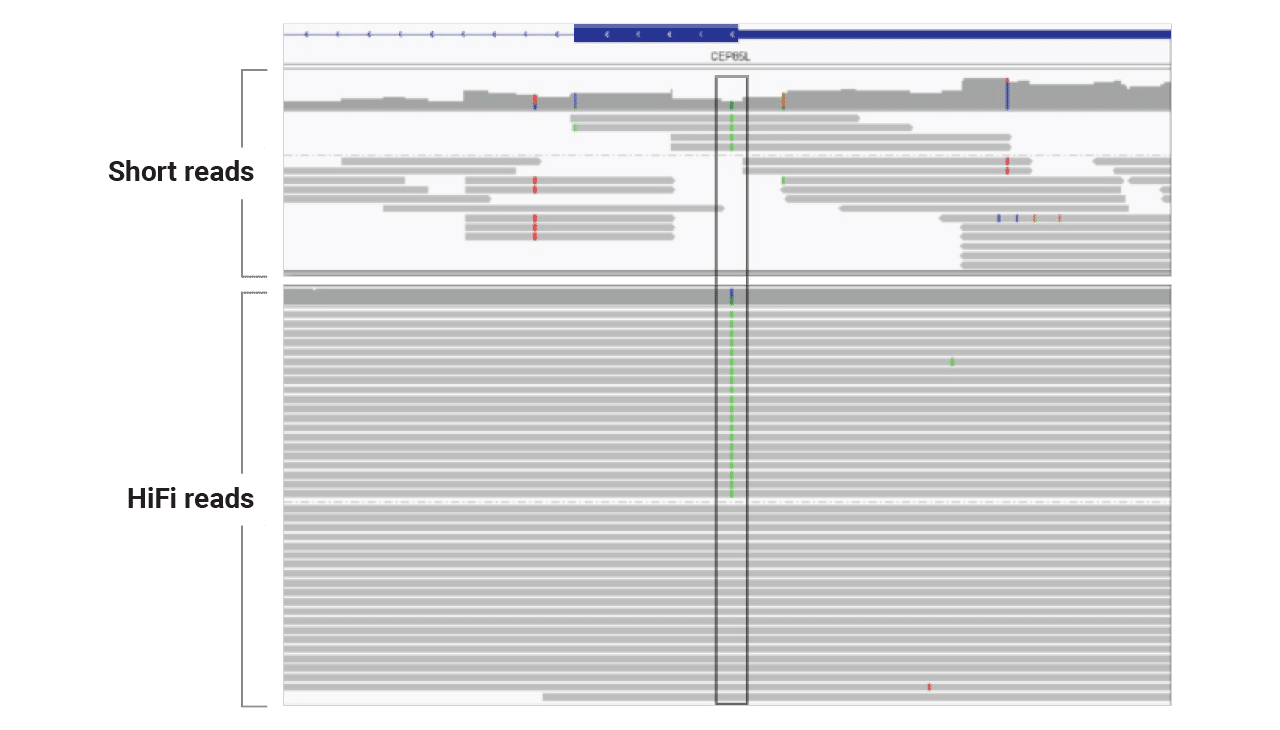
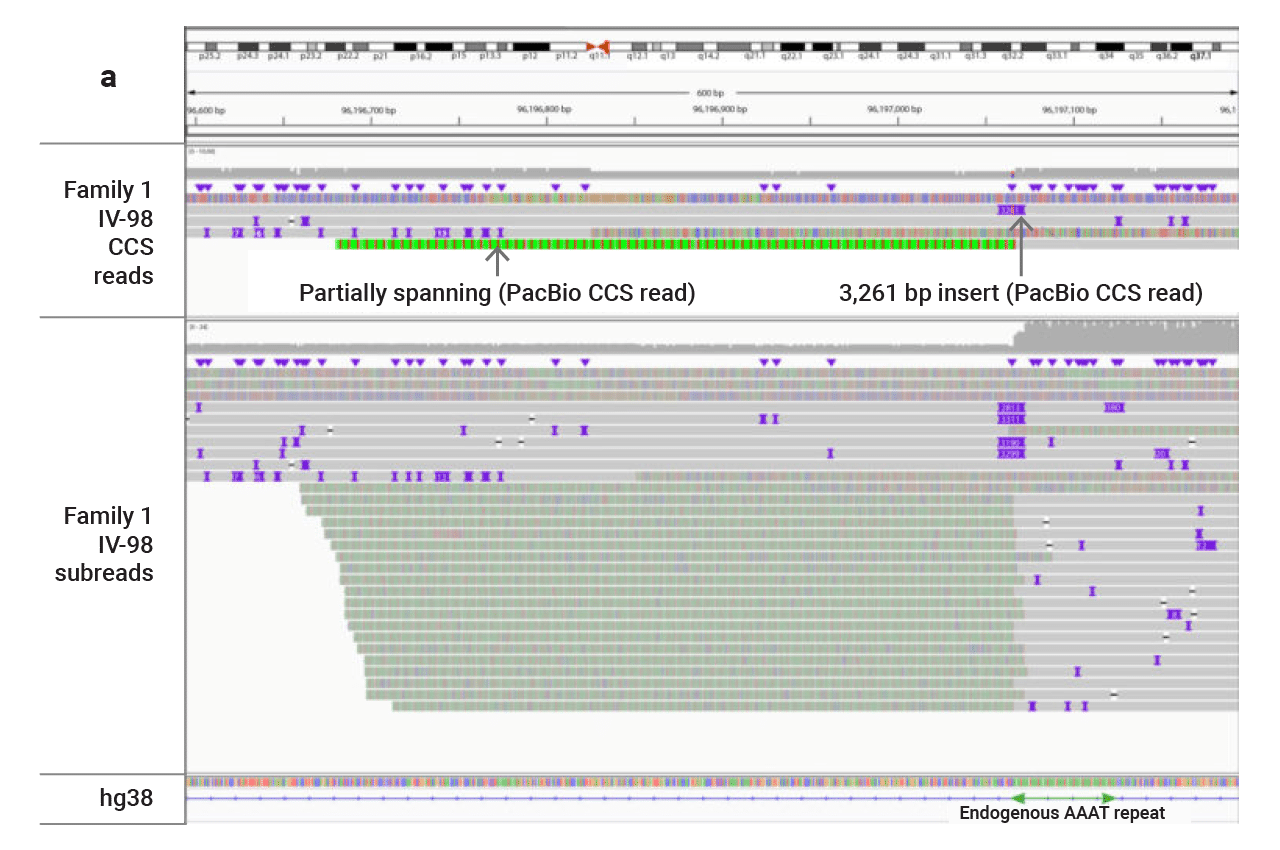
Capturing the full length and sequence of repeat expansions
Repeat expansions have previously been shown to cause a range of diseases and can be tough to characterize accurately with short-read sequencing tools. HiFi sequencing can get through even very long expansions. Recently, scientists from Adelaide Medical School and the Robinson Research Institute linked the expansion of an ATTTC repeat in the first intron of STARD7 with familial adult myoclonic epilepsy.
Repeat expansions are mutations that result in repeating sequence that may extend for hundreds to thousands of bases. For example, the trinucleotide repeat expansion that causes Huntington’s disease, consists of hundreds of CAG repeats.
Phasing rare disease variants across alleles
Phasing involves separating maternally and paternally inherited copies of each chromosome into haplotypes to get a complete picture of genetic variation. Learn more.
Back at Children’s Mercy Kansas City, researchers analyzed the genome of a four-year-old girl with hepatosplenomegaly whose parental genomes were not available. The individual was believed to have Niemann Pick disease type C, but more data was needed to support the theory. HiFi reads showed two key variants located on different alleles of the relevant gene; with the phased variants, scientists were able to confirm the original finding.
The future of rare disease research is bright
Scientists around the world are striving to improve the lives of those affected by rare diseases, translating the latest research approaches and high-quality genomic data into insights that could enable the development of improved diagnostics for rare diseases. As HiFi sequencing continues to shed light on more areas of the genome, it should have a profound effect on our ability to diagnose, understand and ultimately improve treatment for the rare disease community.
To learn more about how PacBio HiFi sequencing is helping advance our understanding of rare disease, watch on-demand presentations from our Virtual Rare Disease Week event or visit our rare disease resource page.
Explore other posts in the Sequencing 101 series
- The evolution of DNA sequencing tools
- Understanding accuracy in DNA sequencing
- Webinar: how long-read sequencing improves access to genetic information
- Introduction to PacBio sequencing and the Sequel II system
- From DNA to discovery — the steps of SMRT sequencing
- DNA extraction — tips, kits, and protocols
- Sequencing 101: ploidy, haplotypes, and phasing — how to get more from your sequencing data
- Why are long reads important for studying viral genomes?
- What’s the value of sequencing full-length RNA transcripts?