It’s well known that finding the genetic cause of rare diseases can be complex — that’s why so many remain unsolved.
But researchers are beginning to get a grasp on just how complex these conditions can be, thanks to the heightened power of PacBio.
PacBio principal scientist Aaron Wenger kicked off a recent webinar with a quote from University of Washington scientist Evan Eichler, who said “there are three key aspects to genetic disease associations: comprehensive variant discovery, accurate allele-frequency determination, and an understanding of the pattern of normal variation and its effect on expression.”
Wenger explained how HiFi reads can address each of these. He noted that while solve rates have improved slightly with each new sequencing technology, more than 50% of cases still remain unsolved. But he added that he is optimistic that the Sequel II System and HiFi reads will be the next transformative technology to increase solve rates, sharing several examples to support his claim.

Kristen Sund (@kristen_sund), a researcher at Cincinnati Children’s Hospital, then shared two specific examples where Single Molecule, Real-Time (SMRT) Sequencing enabled her to untangle complicated structural rearrangements that had eluded earlier detection using other technologies, including short-read sequencing.
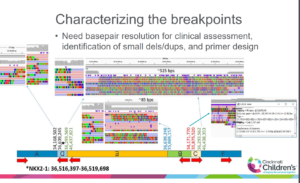
“We set out to understand how new tools like the Sequel may help find an answer for some of our undiagnosed patients,” Sund said. “The goal was to use low-pass long-read SMRT Sequencing to obtain base pair resolution of the breakpoints and identify a gene that was responsible for the phenotype.”
Both cases involved patients with neurological diseases who had a known chromosomal arrangement, but no known gene cause for their phenotype. They both had previous genetic testing with results that were either “normal” or non diagnostic.
The first case, a 9-year-old with short stature and hypotonia, came to the researchers’ attention when she came to the hospital for treatment for nocturnal epilepsy. The girl also had developmental and speech delay, and some self-stimulatory behaviors.
Sund’s testing revealed 31,324 structural variants in the girl’s genome. Notably, one of these structural variants included changes in a protein (MBD5) that regulates gene transcription, and has been linked to developmental delay, speech impairment, seizures, sleep disturbances, and autistic-like behaviors, in a condition known as MBD5-Associated Neurodevelopmental Disorder (MAND).
“I thought we had an answer for this patient, but I still had questions,” Sund said.
So she dug deeper, and discovered the case was far more complex than anyone expected, with not just one straightforward causative variant, but a complicated interplay of inversions, insertions, and deletions. Sund said she is still working through some additional complexity of mapping the exact breakpoint, but she was glad to be able to provide some answers to the girl’s parents.
“This family has been looking for an explanation for their daughter’s epilepsy for nine years. In addition to having an answer for their daughter and resources about the condition and its prognosis, they will also be able to use this information to connect with other families who have a similar diagnosis,” Sund said.
The second case was similarly complex – a movement disorder in a 17-year-old with chorea, myoclonus, anxiety and hypothyroidism, who also had a family history of early death due to lung cancer. In this case, Sund investigated whether it might be a pathogenic variant of the NKX2-1 gene that caused the condition. Paying particular attention to the breakpoints of chromosomal rearrangements, she found many surprises.
“Although this is a small sample size, it does make you wonder whether most – or all – simple rearrangements are actually many chromothripsis events with multiple breaks and rearrangements,” Sund said.
She noted that structural variant analysis should not be limited to coding regions, and concluded that there can be real clinical utility in using long-read technologies for diagnosis, prognosis and treatment of rare disease patients.
“This application will be incredibly meaningful to families who have been searching for a diagnosis,” Sund said.

Sund’s sequencing was carried out at the University of Minnesota Genomics Center, a PacBio Certified Service Provider, as part of the winning 2018 Structural Variation SMRT Grant project.
NGS Operations Manager Archana Deshpande, said: “We have used the Sequel System across a range of applications, including whole genome sequencing for plants, Iso-Seq analysis of gene expression, multiplexed microbial whole-genome sequencing, and the detection of structural variants. Our new Sequel II System promises even more and we are extremely pleased with the amount and quality of data our preliminary runs have generated.”
Watch the webinar:
See additional information and examples of the use of SMRT Sequencing in rare disease research:
The Pathologist: Solving Rare Disease with SMRT Sequencing
A Rare Opportunity to Help Tackle Daughter’s Rare Disease
Review: Long-Read Sequencing Helps Uncover Genetic Basis for Rare Disease
SOLVE-RD Team Adopts PacBio Sequel II System to Solve Rare Diseases