No. PacBio does not accept samples for sequencing. See the list of our global certified service providers (CSPs) by geographic region.
Support FAQs
Email support@pacb.com with the name of your employer or institution and your address and we will connect you to the relevant team for your organization.
You can find additional information on our training where you can search for and download online webinars, e-modules, and other training documents. You can also find various applications (WGS, targeted, epigenetics, RNA and metagenomics) on our sequencing methods page. If you have any questions about our training materials, contact us.
Here is an overview of HiFi applications
You can find the protocols (procedure & checklists) on the protocols page (filtered view from featured documentation).
The only supported sample type is human blood or cell line extracted with PanDNA or CBB kits. We have seen success internally and at customer sites using other human tissues and extraction kits. Please see our PureTarget app note for more details.
When using a sample type other than Nanobind-extracted human blood or cell line we recommend the following:
- First, demonstrate success using supported sample types, starting with an 8-plex and increasing sample quantity thereafter.
- Introduce new sample types or extraction methods in limited numbers, for example, 3 or fewer new sample types in an 8-plex of otherwise controls.
- See table 3 in the app note for more information about samples that are officially supported, have been tested, or are not recommended.
Non-human samples are untested and not supported as the guide RNAs used in the panel are designed for the human genome. With non-human samples, you can follow our suggestions on custom guide design. The region captured in the panel is available in Panel coordinate (XLSX).
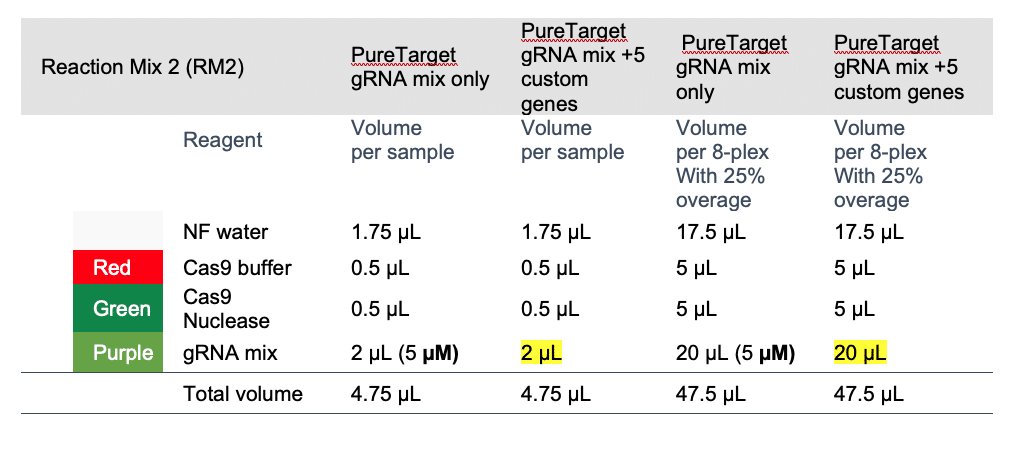
The PureTarget 20 gene panel is at 5 µM and includes 20 pairs of gRNAs made up of 40 individual gRNAs at 0.125 µM each.
To add on custom genes to the panel, for simplicity, we recommend the following:
- Prepare your custom gRNAs pair per gene at final 5 µM.
- Spike into the PureTarget gRNA mix to prepare custom gRNA mixes prior to making ‘Reaction Mix 2 (RM2)’ in step 3 of the Procedure & checklist – Generating PureTarget repeat expansion panel libraries, while ensuring the total sum molarity of the final mix is 5 µM; the volumes of gRNAs you mix should be proportional to the number of guide pairs, for example add 2 µL of a 1-plex mix to 40 µL of the 20-plex PureTarget panel to keep the sum final molarity at 5 µM.
- Do not change any of the master mix volumes in the protocol.
Please consult with PacBio field application scientist or global service and support for more specific details on library prep
Custom PureParget panels are not supported by PacBio
We can to users interested in designing and optimizing their own panels. In all cases, we recommend first demonstrating success on the PureTarget repeat expansion panel using supported sample types before adding new guide RNAs or testing a custom set of guides.
Adding more validated repeat expansion targets: This “flavor” of customization is relatively low risk. Internally, we have demonstrated success in adding up to 5 pairs of guides for additional repeat expansion targets. The design strategy is the same as the fixed 20 gene panel: design guides ~2 kb upstream and downstream of the repeat expansion region with fragments ranging from 3-5 kb.
Adding non-repeat expansion targets: Internally, we have demonstrated success in adding a pair of guides to target a full-length gene with fragment size of ~5 kb, similar in size to the PureTarget repeat expansion panel. Please consult with your field application bioinformatics support scientist for the recommended variant calling tools for your targets.
All other designs like capturing fragments longer than 5 kb or using a tiled design are untested and we do not have any recommendations on design or optimization.
A software tool we recommend for designing guide RNAs is Custom Alt-R™ CRISPR-Cas9 guide RNA from Integrated DNA Technologies (IDT) available here: https://www.idtdna.com/site/order/oligoentry/index/crisprcustom.
We recommend ordering SygRNAs (Synthetic Single Guide RNAs) from MilliporeSigma or sgRNA from IDT rather than the two-part targeting system composed of a separate tracrRNA and crRNA. This way, you only need to order one gRNA and the preparation is simplified (there is no annealing of the two molecules).
PacBio has developed automation methods for extraction on the Thermo Fisher Kingfisher Apex, Flex, and Duo. PacBio has also developed methods for size selection using SRE, shearing with pipette tips, and bead cleanup on the Hamilton Microlab Prep. You can find all of these methods at the third-party equipment webpage. For methods on any other equipment, please reach out to the support team of the appropriate vendor.
Please use our automation workflow selector tool to determine which of our PacBio Compatible automation solutions are best for your application.
You can find all the resources for barcoding all types of samples on the multiplexing page
- For genomic or metagenomic DNA, see page 1 of the following: Procedure & checklist – Preparing whole genome and metagenome libraries using SMRTbell prep kit 3.0
- For 16S, see page 7 of the following: Procedure & checklist – Preparing Kinnex libraries from 16S rRNA amplicons
- For other amplicons, see page 3 of the following: Procedure & checklist – Preparing multiplexed amplicon libraries using SMRTbell prep kit 3.0
- For bulk mRNA, see page 1 of the following: Procedure & checklist – Preparing Kinnex libraries using the Kinnex full-length RNA kit
- For single-cell cDNA, see page 1 of the following: Procedure & checklist – Preparing Kinnex libraries using Kinnex single-cell RNA kit
Analysis pipelines are enabled for many applications using SMRT Link.
In addition, a large assortment of command line computational tools and workflows that cover many different types of analyses can be found on the PacBio Github repository.
An overview of HiFi applications and associated analysis tools is also available.
PacBio software is provided for free from our software + bioinformatics webpage, Bioconda, DockerHub, Quay.io, and the PacBio Github. Depending on the type of analysis you need to do, it may be easier to install our SMRT Link software suite or install particular computational tools or workflows to your Linux server or Cloud instance. Please reach out to technical support or Field Applications Bioinformatics Scientists for more information.
HiFi reads are produced using circular consensus sequencing (CCS) mode on PacBio long-read systems. HiFi reads provide base-level resolution with 99.9% single-molecule read accuracy.
HiFi reads can be used across a wide range of SMRT sequencing applications, from whole genome sequencing for de novo assembly, comprehensive variant detection, epigenetic characterization, RNA sequencing and more.
With reads tens of kilobases in length you can readily assemble complete genomes and sequence full-length transcripts. HiFi sequencing provides exceptional read lengths without compromising throughput or accuracy. For more information visit the How does HiFi sequencing work? or HiFi sequencing technology page.