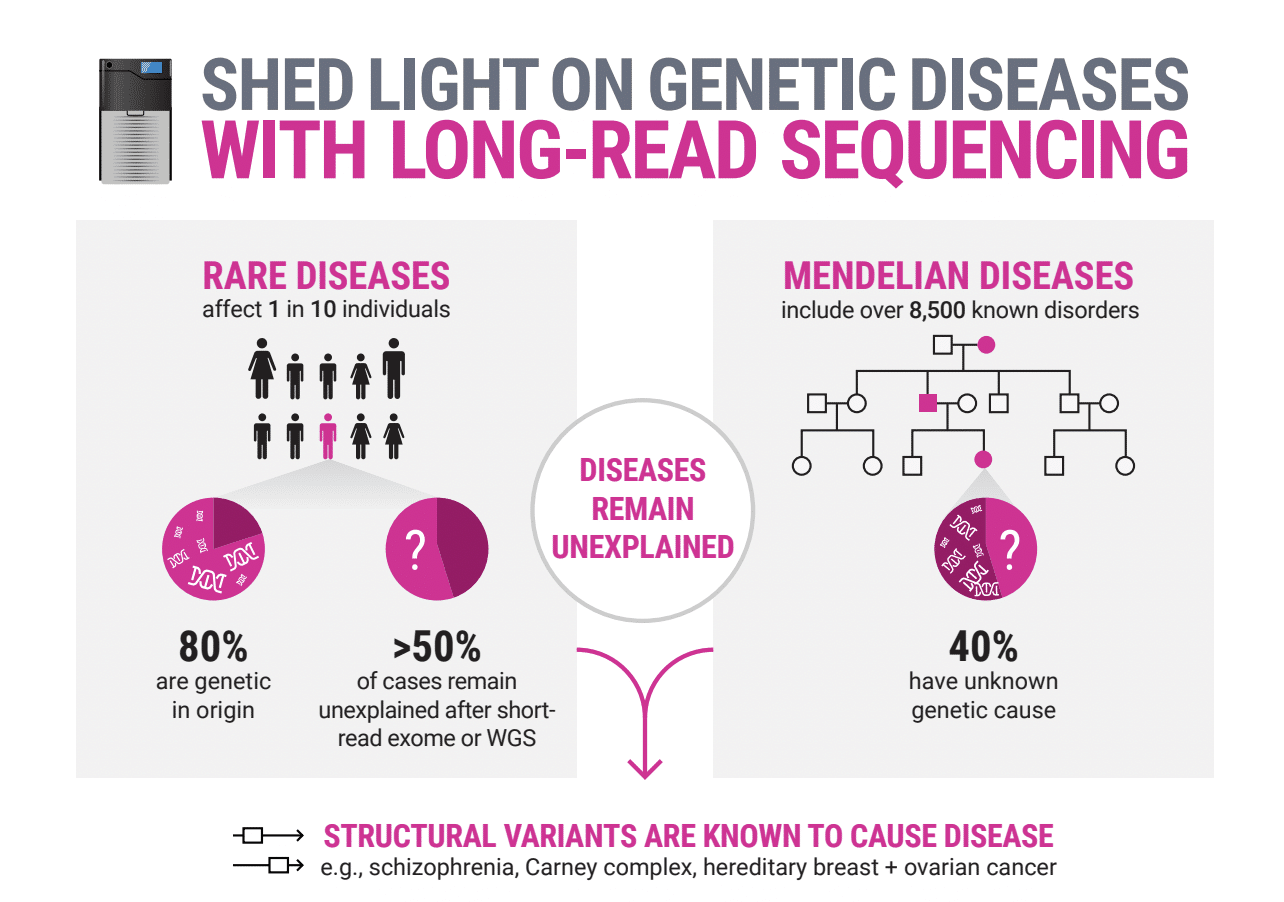
Rare diseases need better solutions
Although rare diseases are individually rare, collectively they are common, affecting 1 out of 15 people worldwide with an estimated >70% of cases being genetic in origin and >50% of cases remaining unsolved. This is despite many people undergoing diagnostic tests using approaches including microarrays, whole exome, or short-read whole genome sequencing.
Highly accurate long-read sequencing, known as HiFi sequencing, provides a higher resolution approach compared to previously used technologies to better understand the genetic causes of rare disease.
Repeat expansion panel for clinical research
The PureTarget repeat expansion panel is a kitted solution enabling testing laboratories to more comprehensively genotype 20 genes associated with serious neurological disorders – including difficult-to-sequence genes with tandem repeat expansions. The panel offers a robust, end-to-end workflow designed to reduce iterative testing using legacy technology and reduce the time needed to identify neurological disease-causing variants and associated methylation signatures.
Rare disease research. Can HiFi sequencing help shorten the journey to explanations?
A team of researchers at Radboud University Medical Center (Radboudumc) in Nijmegen, Netherlands is looking at how they can reduce the time it takes to identify and name a person’s rare genetic disease. Team leader, Lisenka Vissers shares her vision for rare disease and how HiFi sequencing can help Radboudumc get there.
HiFi sequencing for rare diseases
With accuracy 99.9% and long read lengths up to 25 kb, HiFi sequencing allows scientists to find probable causative pathogenic variants and identify novel disease-associated genes with:
- Best-in-class variant calling for more variant types including single nucleotide variants (SNV), indels, copy number variants (CNVs), and structural variants (SVs)
- More complete, accurate, and phased assemblies of the human genome — including regions previously inaccessible to other technologies — for accurate typing of genes including HLA and CYP2D6
Generate new insights by unlocking tandem repeats in the human genome not explicitly profiled by common whole genome analysis pipelines.
Tandem repeats are one of the most abundant types of variation in the human genome, and due to their repetitive nature, they have the highest mutation rate in the genome. This genomic instability is a major driver of disease. There are more than 50 disorders caused by expansions of short tandem repeats (STRs), and several variable number tandem repeats (VNTRs) have been shown to be involved in common complex disease like Alzheimer’s, autism, epilepsy, ALS, and others. Although tandem repeats play a key role in human health and disease, they are commonly not profiled as part of whole genome analysis pipelines.
This is not because they are considered unimportant, but because for so long it has been impossible to sequence them using short-read sequencing, directly impacting analysis development. As part of the recent gapless human genome sequencing efforts, it has been estimated that 50–80% of structural variants are actually tandem repeats.8 However, since the current analysis pipelines are not optimized for tandem repeats, they are often either reported as indels if they are smaller or as clusters of variants if they are larger. HiFi sequencing enables a more complete and accurate calling of repeat expansions and identification of medically relevant interruption sequences along with methylation profiles.
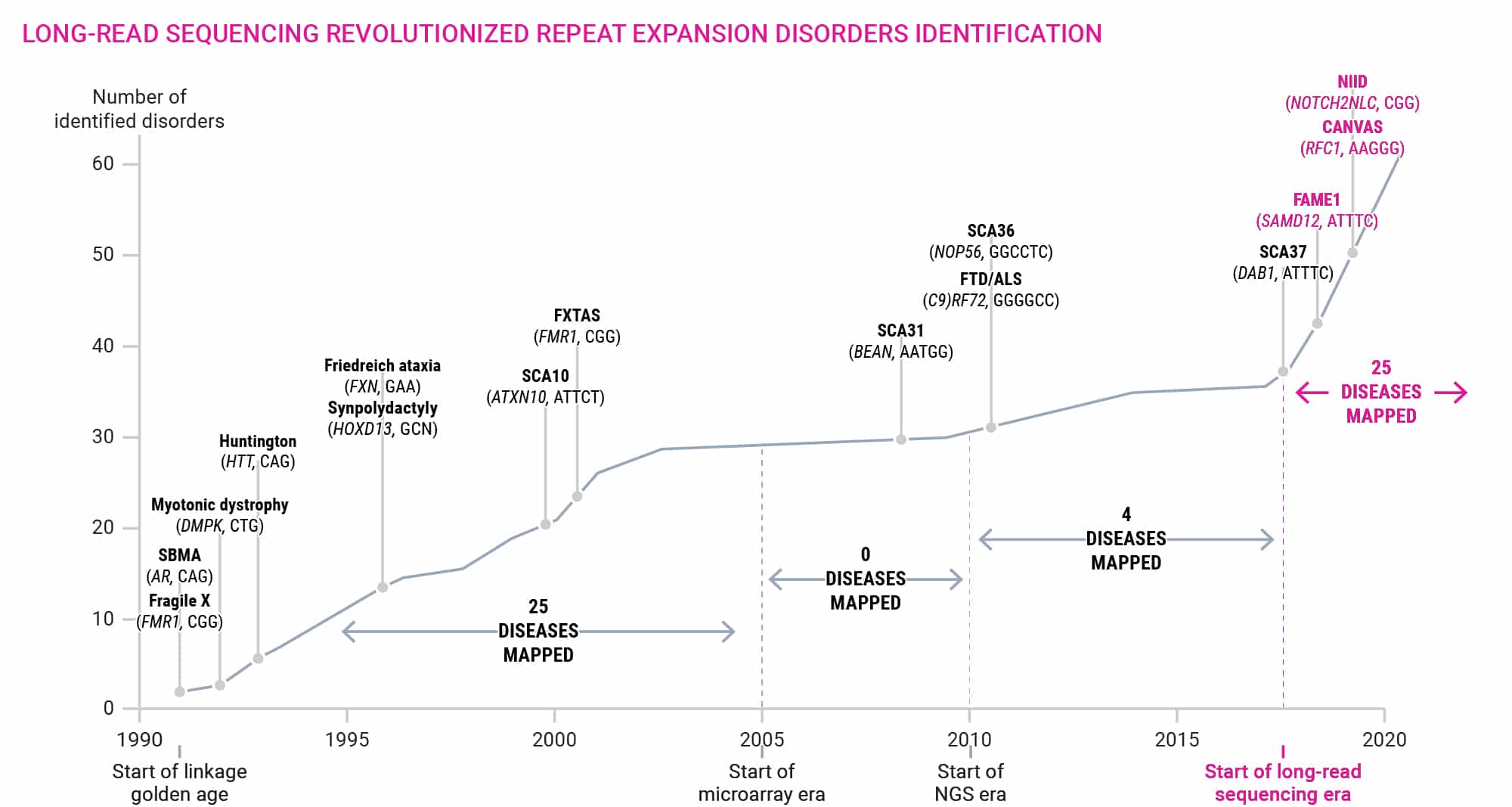
Figure 8. Timeline of repeat expansion discovery in human disorders.9
8Estimated by Adam English (Baylor College of Medicine) with Truvari software; SV from 36 haplotype-resolved assemblies spanning at least 50 bp; TR definitions from the UCSC SimpleRepeats track
9Depienne et. al (2021) 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges? AJHG,108(5): 764-785
Genomic Answers For Kids
Spotlight: Children's Mercy Hospital uses HiFi sequencing in Genomic Answers for Kids study
Children’s Mercy Kansas City has expanded its investment in PacBio Sequel IIe systems to scale up whole genome research initiatives focused on rare disease research, and the results are already beginning to add up. Read about their success in resolving the mysteries of rare and inherited disease in children in our newly released customer success story.

Whitepaper
Bulk and single-cell isoform sequencing for human disease research
In this whitepaper, we discuss the features of bulk and single-cell isoform sequencing with the Iso-Seq method and their applications in human genomics.
Leveraging IsoForm-Level RNA Sequencing to Understand Rare Disease Pathogenesis
In a next-level exploration of the latest advancements in RNA sequencing technology and its application in the study of rare and inherited diseases, hear from Dr. Carolina Jaramillo Oquendo, a Research Fellow at the University of Southampton, as she presents her analyses of rare disease samples, in collaboration with the University Hospital Southampton NHS Foundation Trust.
Spotlight
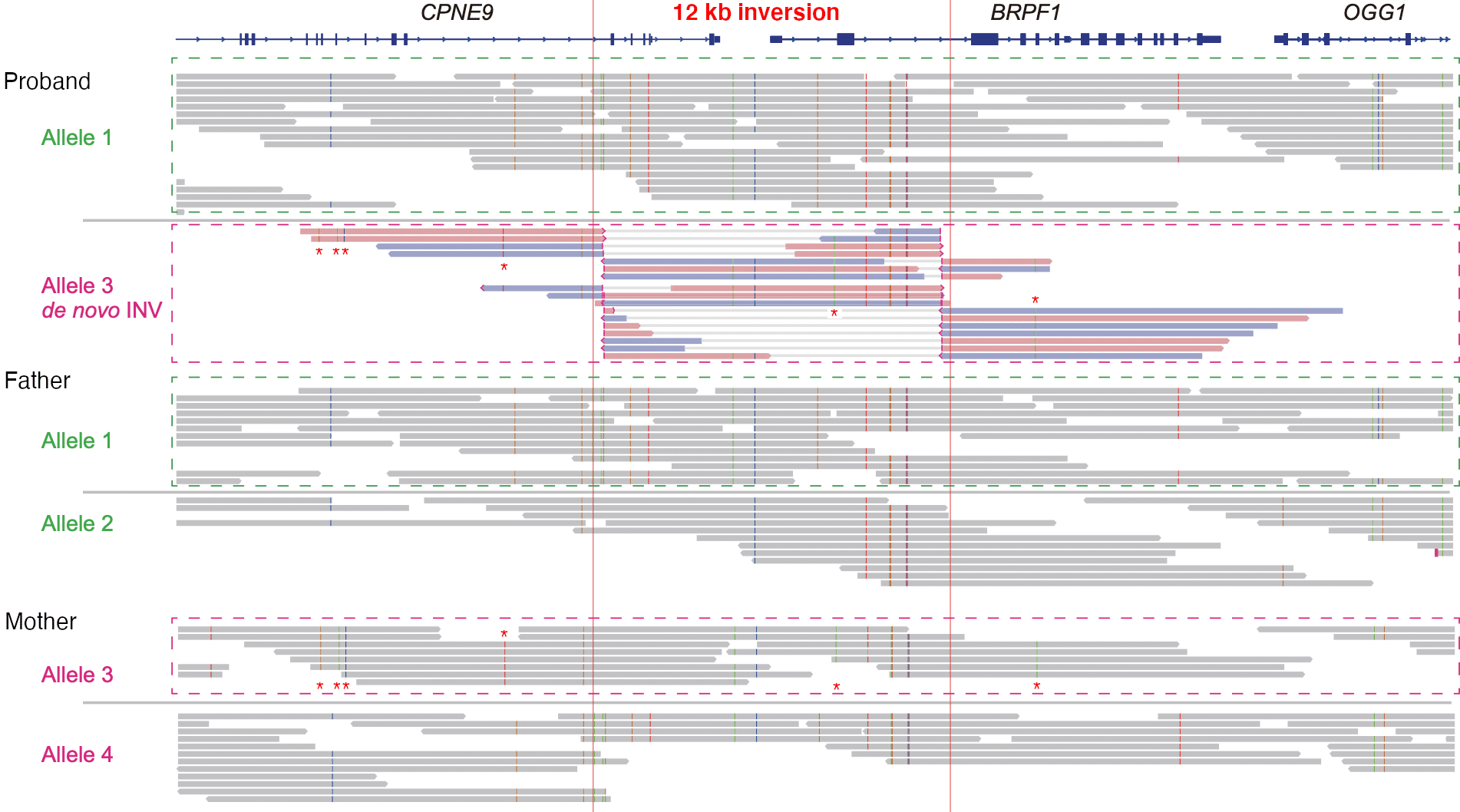
12 kb inversion identified as probable causative effect in syndromic intellectual disability
HiFi sequencing of a trio identifies a pathogenic heterozygous 12 kb de novo inversion that disrupts the BRPF1 gene. SNVs (marked with “*”) show that the inversion occurred on the maternal allele #3. Read more about this study in the blog post.
Mizuguchi, T., et al. (2021) Pathogenic 12-kb copy-neutral inversion in syndromic intellectual disability identified by high-fidelity long-read sequencing. d. 113(1).
Explore the range of HiFi sequencing applications
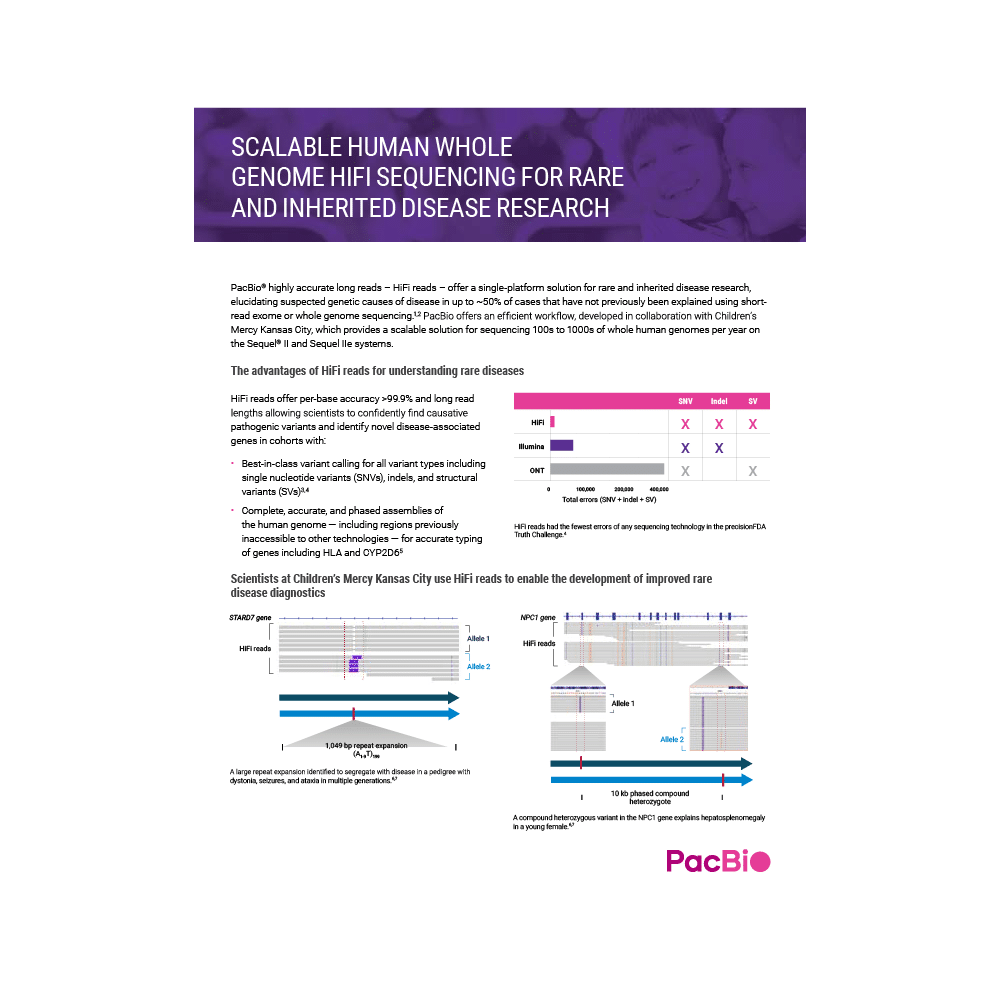
Scalable human whole genome HiFi sequencing for rare and inherited disease research
Explore our efficient and scalable workflow, developed in collaboration with Children’s Mercy Kansas City, for high-throughput sequencing and comprehensive variant detection to better understand the probable genetic causes of rare and inherited diseases.