Advance your microbial genomics research with HiFi sequencing
HiFi sequencing on the PacBio systems provides a deeper understanding of microbes and microbial communities by giving scientists the ability to:
- Resolve complete microbial genomes and detail their mobile elements
- Characterize epigenetic modifications and their impact on critical bacterial traits
- Investigate viral pathogenicity, evolution, and selection
- Understand microbial communities
Application brief
Microbial genomics at scale with PacBio HiFi sequencing
Microbial genomics with PacBio long-read HiFi sequencing is accurate, scalable, and cost effective. Automated end-to-end workflows from extraction to analysis and optimized kits drive greater throughput for HiFi microbial sequencing at lower cost.
Spotlight
MINING STREPTOMYCES FOR ANTIMICROBIAL GOLD
The genome of a novel Streptomyces anulatus strain isolated from a Chinese medicinal herb was sequenced with PacBio and fully assembled to reveal 52 putative biosynthetic gene clusters, including numerous with significant differences from known clusters. Growth under diverse conditions produced two novel compounds, with more potential yet to be unlocked.
Liu, et. al. (2022). Exploration of diverse secondary metabolites from Streptomyces sp. YINM00001, using genome mining and one strain many compounds approach. Front Microbiol.
Application brief
MICROBIAL WHOLE GENOME SEQUENCING — BEST PRACTICES
- Generate closed chromosomes and plasmids from even the most repeat-dense and GC-rich genomes easily, affordably, and at high throughput
- Identify ever-evolving genes associated with toxicity, virulence, and antimicrobial resistance
- Precisely identify strains, serotypes, and plasmids to track pathogen outbreaks in human, plant, and animal species, through food systems, hospitals, and communities
- Comprehensively characterize microbes to facilitate scientific breakthroughs and discovery
Blog
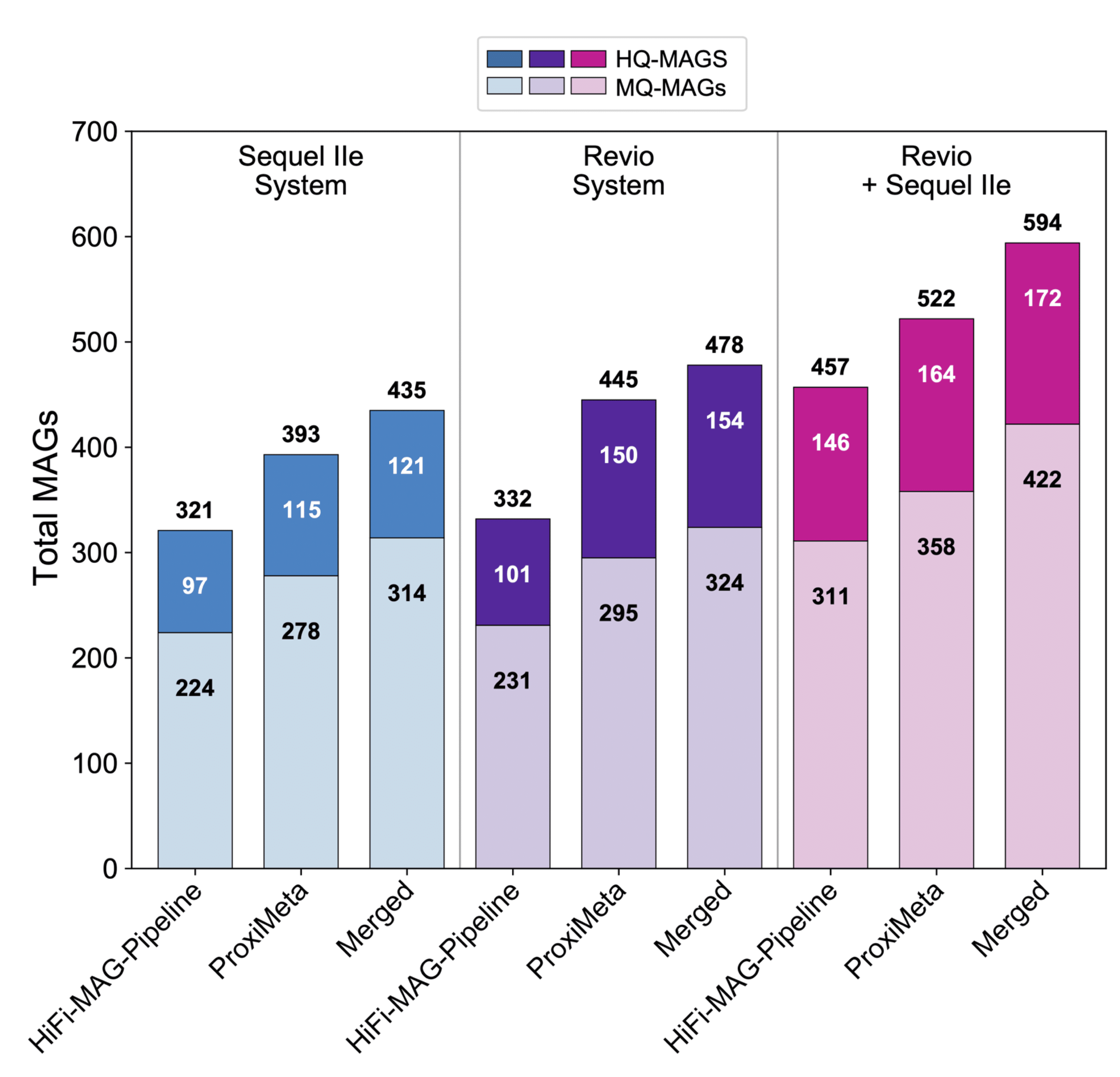
REVIO METAGENOMICS + KINNEX 16S DATA RELEASE DEMONSTRATE FUTURISTIC MICROBIOME CAPABILITIES
Microbiology forms the bedrock of modern genetics and biotechnology. Whether its basic science, alternative energy development, drug discovery, or human health – our planet’s smallest organisms play a critical role in the bigger picture. Even so, most microbial life remains unknown to science. And that’s why cutting-edge microbial ecology and metagenomics research seeks to reveal the biological secrets of this hidden majority. How do we go about understanding what we have thus far struggled to observe in detail? Sequencing. Really good sequencing.
Blog
Meeting the threat of antimicrobial resistance with HiFi sequencing and artificial intelligence
After conducting a battery of technology comparisons, OpGen’s Ares Genetics has adopted PacBio HiFi microbial whole genome sequencing. The company believes the read length and high accuracy of HiFi data will enable it to develop a suite of cutting-edge AI solutions with potential to revolutionize the way we combat the spread of antimicrobial resistance.
Solutions Guide
A new standard in pathogen genomics
Detect, track, and characterize pathogens to a new standard of completeness and precision. PacBio solutions offer exceptional accuracy and long read lengths to support a wide range of pathogen surveillance applications.
Spotlight
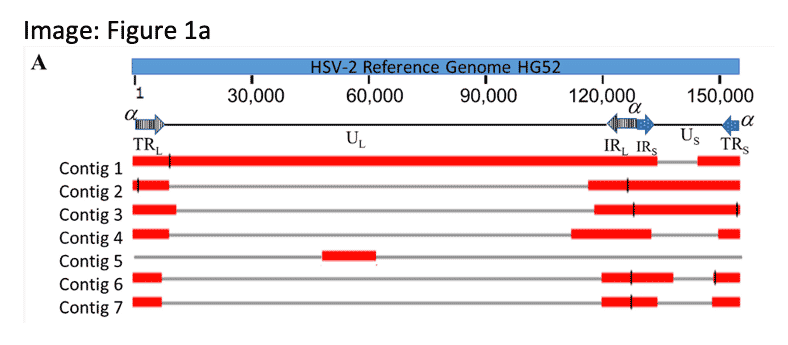
ADVANCING CLINICAL RESEARCH WITH BETTER VIRAL GENOMES
Despite its importance as a clinical target, only 4 herpes simplex virus type 2 strains have complete genomes due to dense repeats and ~70% GC content. Here, scientists report the complete de novo assembly and annotation of an additional strain commonly used in research, HSV-2 strain G.
Chang, W. et. al. (2022). Complete genome sequence of herpes simplex virus 2 strain G. Viruses 14(3), 536; https://doi.org/10.3390/v14030536.
Blog
Cost-effective microbiome and metagenomic HiFi sequencing with PacBio service providers
Full-length 16S and shotgun metagenome HiFi sequencing with certified PacBio service providers can deliver remarkably affordable microbiome and metagenomic data with explanatory power that is second to none.