Explore microbiome + metagenomics with confidence
Highly accurate long reads – HiFi reads – with single-molecule resolution are ideal for full-length 16S rRNA sequencing, shotgun metagenomic profiling, and metagenome assembly, so that you can:
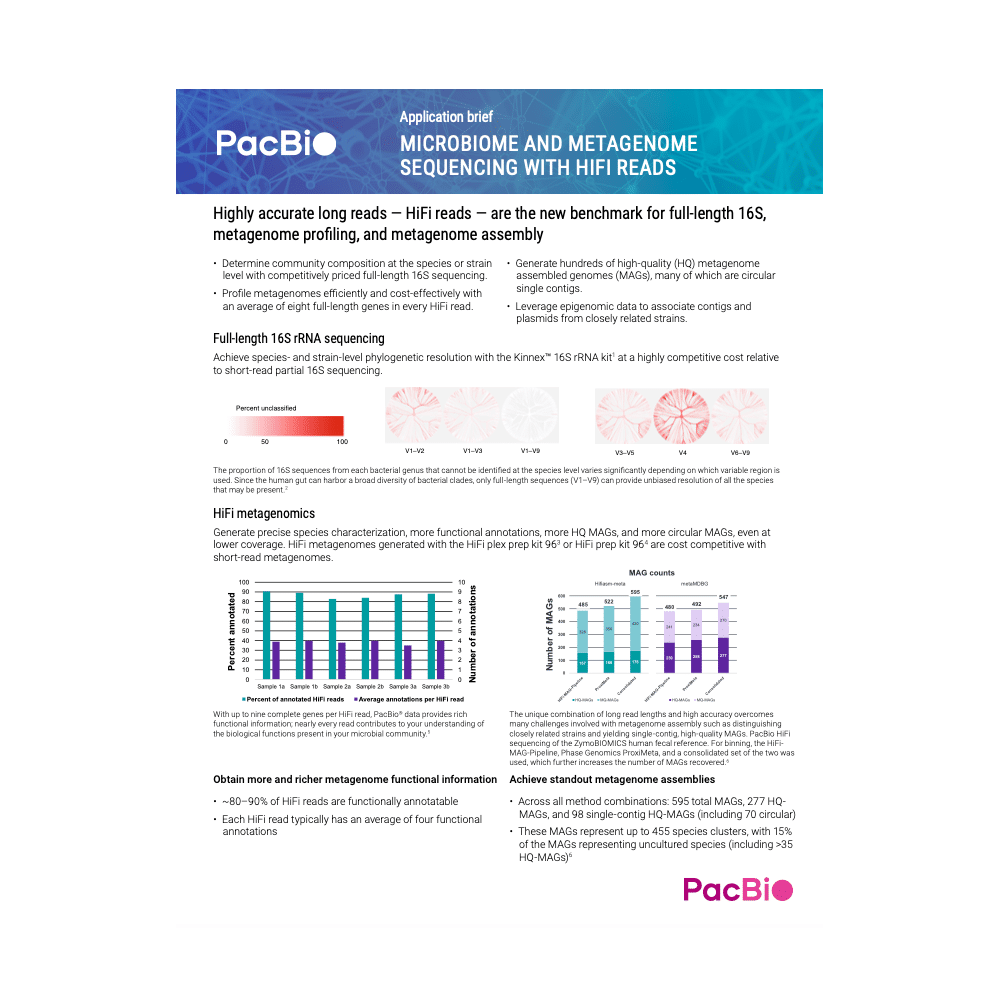
- Determine community composition at the species or strain-level with competitively priced full-length 16S sequencing
- Identify 6-8 full-length genes in every HiFi read with efficient, cost-effective metagenomic profiling
- Generate hundreds of high-quality (HQ) metagenome assembled genomes (MAGs), many of which are single contig, circular MAGs
- Leverage epigenomic data to associate contigs and plasmids from closely related strains
Suggested microbiome + metagenomics sequencing products
Microbial sequencing methods
HiFi metagenomics sees performance enhancements across the board
Shotgun metagenomics with PacBio highly accurate long-read sequencing (aka HiFi sequencing) has been the premier approach for generating microbial datasets that are both very high quality and stunningly complete. Now, the Revio system offers faster and more cost-effective long-read sequencing for metagenomics researchers, allowing for more high-quality and complete microbial datasets and access to new frontiers of discovery.
Revio metagenomics + Kinnex 16S data release demonstrate futuristic microbiome capabilities
PacBio has changed the game again in metagenomics with the release of the Revio system. And now, PacBio is pleased to present the Kinnex 16S rRNA kit for PacBio long-read sequencing platforms. This exciting new kitted solution enables microbiologists to boost 16S taxonomic data generation by nearly an order of magnitude!
Check out some results and download some data to try out for yourself.
Microbiome + Metagenomics Datasets
Blog
Can microbial communities turn plastic waste into sustainable food?
Before being awarded a PacBio SMRT Grant in 2021, the researchers only had short-read data on the microbial communities used to digest and flexibly convert plastics. This left the team with an incomplete picture of the composition and collective functional capabilities of their bacterial consortia. So, to fill this information gap and advance their process design they utilized PacBio long-read metagenomics to successfully capture, and close metagenome-assembled genomes for strains found within their experimental microbial communities.
Notario, E., et al. (2023). Amplicon-Based Microbiome Profiling: From Second- to Third-Generation Sequencing for Higher Taxonomic Resolution. Genes, 14(8), 1567. https://pubmed.ncbi.nlm.nih.gov/37628619/
Spotlight
Why guess with partial 16S sequencing when full-length 16S is the best?
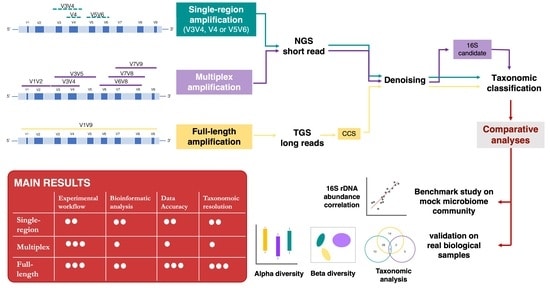
This study compared three different methods to achieve the deepest microbiome taxonomic characterization: (a) the single-region approach, (b) the multiplex approach, covering several regions of the target gene/region, both based on NGS short reads, and (c) the full-length approach, which analyzes the whole length of the target gene thanks to TGS long reads [PacBio]. Analyses carried out on benchmark microbiome samples, with a known taxonomic composition, highlighted a different classification performance, strongly associated with the type of hypervariable regions and the coverage of the target gene. Indeed, the full-length approach showed the greatest discriminating power, up to species level, also on complex real samples. This study supports the transition from NGS to TGS [PacBio] for the study of the microbiome.
Application note
Full-length HiFi sequencing of eukaryotic ITS and rRNA amplicons
PacBio HiFi long-read amplicon sequencing has demonstrated advantages for high-throughput, high- resolution eukaryotic barcode sequencing.
Blog
Kinnex kits enable full-length 16S and RNA sequencing at scale
When new sequencing instruments are released, great leaps in genomics capabilities follow. But did you know that for some applications like 16S microbiome research, gene annotation, or cancer transcriptomics, “new-instrument-level” performance can be achieved with something as simple as an off-the-shelf kit?
On-demand webinar
METAGENOME ASSEMBLY AND CHARACTERIZATION OF A POOLED HUMAN FECAL REFERENCE
Leading scientists from PacBio and Zymo Research present state-of-the art HiFi metagenomic sequencing solutions that enable microbiome researchers to make important discoveries.
Brochure
SETTING NEW STANDARDS IN METAGENOMICS AND MICROBIOME SEQUENCING
Accurate and flexible HiFi solutions for metagenomics and microbiome sequencing allow researchers to study microbes and microbial communities in high resolution without the need for culturing. The exceptional accuracy and long read lengths of HiFi sequencing gives scientists the flexibility and high resolution they need.
Spotlight
Both read length and accuracy matter for metagenome assembly and functional profiling
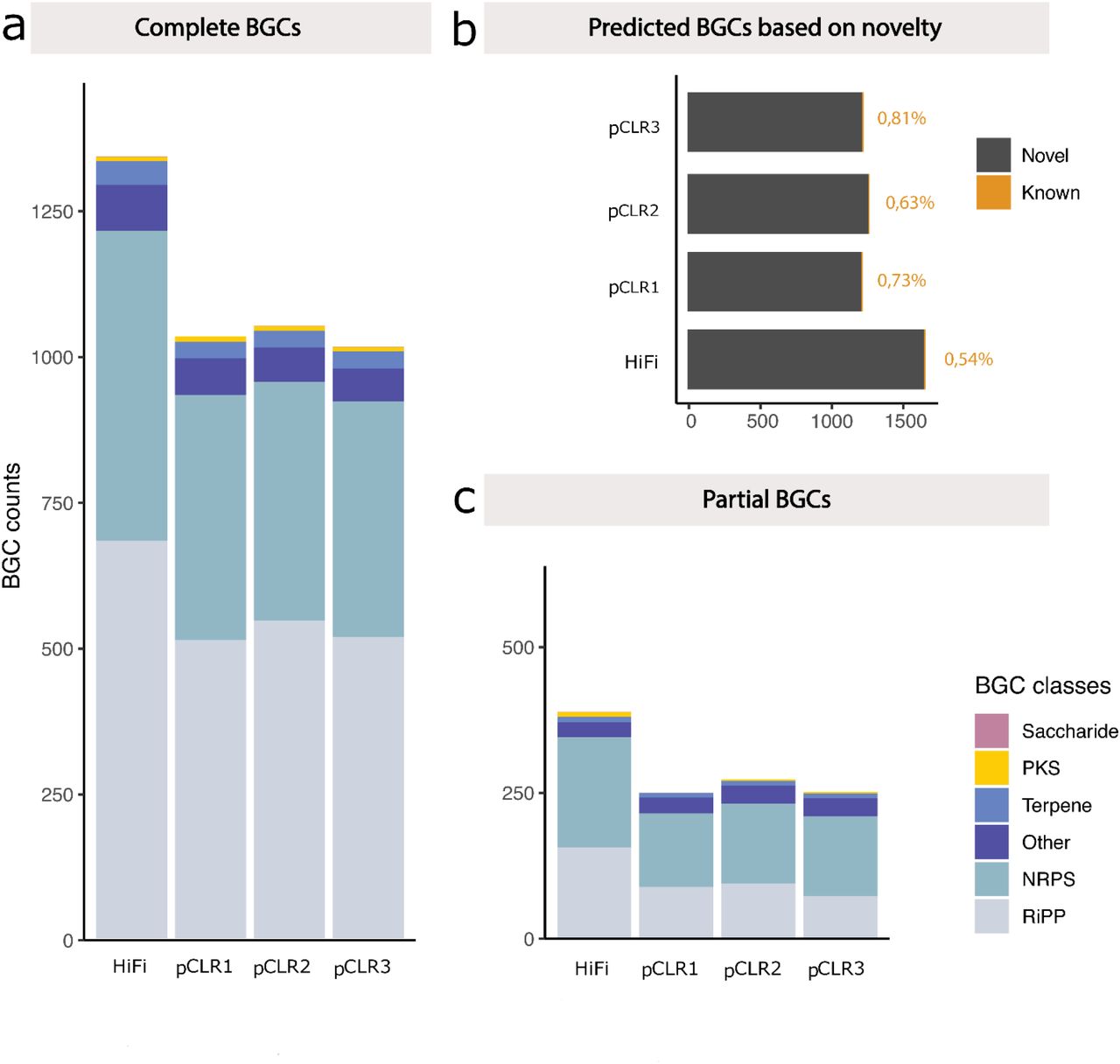
Comparing analysis results using HiFi data from a sheep fecal sample to results using lower accuracy subreads from the same data set show the impact of improving accuracy on assembly contiguity, MAG recovery and gene discovery. In the figure, the more contiguous HiFi assembly enabled recovery of 25% more complete biosynthetic gene clusters (BCGs) than noisy CLR long reads. Explore this research further:
Bickhart, D.M. et al. (2022). Generation of lineage-resolved, complete metagenome-assembled genomes from complex microbial communities.. Nature Biotechnology, 0.1038/s41587-021-01130-z
Library preparation
Workflow: from DNA to resolved microbial communities
Prepare targeted sequences or shotgun libraries to characterize your microbial community.
| Samples and reads/data per SMRT Cell* and cost estimate† | ||||||||||||||
| Application | Protocol / Library prep | Sequel II / IIe SMRT Cell 8M | Revio SMRT Cell 25M | Data analysis tools | ||||||||||
| Full-length 16S rRNA: high-resolution, cost-effective screening for microbial community studies | Amplification of bacterial full-length 16S gene with barcoded primers | Preparing Kinnex libraries from 16S rRNA amplicons |
Standard 16S: |
Kinnex 16S: |
Standard 16S: |
Kinnex 16S: |
Analyze full-length 16S data with either DADA2, QIIME 2, microbiomehelper, OneCodex, EZBiome, or with the PacBio GitHub pipeline |
|||||||
| Metagenome profiling: Unbiased compositional and functional characterization of microbial communities‡ |
Create ~10 - 15 kb libraries for HiFi metagenome sequencing |
48 communities |
96 communities |
Study metagenome taxa and functions using DIAMOND and MEGAN-LR or Sourmash with the PacBio GitHub pipeline or with BugSeq (a PacBio Compatible Partner) |
||||||||||
| Metagenome assembly: Generation of complete or near-complete metagenome-assembled genomes (MAGs) from microbial populations |
Create ~10 - 15 kb libraries for HiFi metagenome sequencing |
4 communities |
12 communities |
De novo assemble high-quality metagenomes with hifiasm-meta, metaFlye, or metaMDBG and perform QC, binning, classification, and evaluation with the PacBio GitHub pipeline |
||||||||||
|
* Read lengths, reads/data, and number of samples per SMRT Cell and other sequencing performance results vary based on sample quality/type and insert size. † Prices, listed in USD, are approximate and may vary by region. Pricing includes library and sequencing reagents run on a Sequel II / IIe or Revio system and does not include instrument amortization, other reagents, or DNA extraction. ‡Metagenomic DNA can be extracted using commercially available kits from suppliers such as Zymo Research and Qiagen. For metagenome sequencing libraries from sample types that may contain inhibitors or contaminants, it is recommended to further clean up the extracted DNA with a column-based cleanup such as the DNeasy PowerClean Pro Cleanup Kit (Cat. No. 12997-50). |
||||||||||||||
Explore recommendations for all SMRT sequencing applications
Extract DNA using the commercially available kit of your choice
Application brief
Microbiome and metagenome sequencing with HiFi reads
Highly accurate long reads – HiFi reads – with single-molecule resolution make Single
Molecule, Real-Time (SMRT) sequencing ideal for full-length 16S rRNA sequencing,
metagenome profiling, and metagenome assembly.
BUGSEQ JOINS PACBIO COMPATIBLE PRODUCTS, ENABLES LEADING ACCURACY INFECTIOUS DISEASE ANALYSIS FOR LONG READS
BugSeq Bioinformatics, a leading provider of infectious disease bioinformatic solutions, has joined the PacBio Compatible program as a PacBio partner. Combined with the high quality, long reads of the PacBio HiFi platform, BugSeq enables recovery of complete microbial genomes, strain-level profiling, and detection of antimicrobial resistance mutations from metagenomic samples.
Application brief
BugSeq taxonomic and functional profiling with HiFi metagenomics
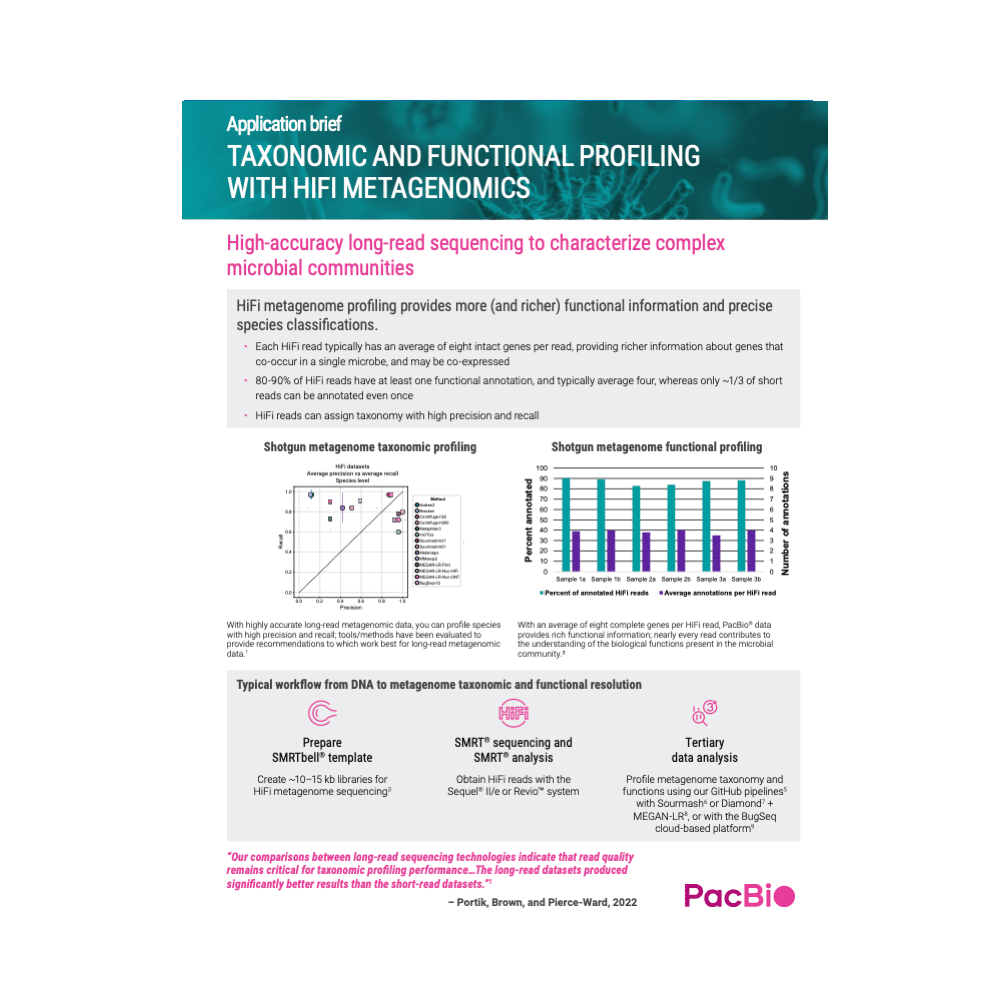
HiFi metagenome profiling provides more (and richer) functional information and precise species classifications.
- Each HiFi read typically has an average of eight intact genes per read, providing richer information about genes that co-occur in a single microbe, and may be co-expressed
- 80-90% of HiFi reads have at least one functional annotation, and typically average four, whereas only ~1/3 of short reads can be annotated even once
- HiFi reads can assign taxonomy with high precision and recall
Microbiome blog
Digest HiFi microbiome and metagenomics research with Publications at a Glance
If you are interested in seeing how PacBio technology is making an impact in current metagenomics and microbiome research, our new Publications at a Glance are an effortless and convenient way to access the latest PacBio-driven discoveries from the community.
This first Publications at a Glance series is comprised of a single-page document (written by PacBio scientists) that delivers busy researchers a quick and easy-to-digest summary of important peer-reviewed articles in microbiome and metagenomics research. Each one highlights key findings from the featured publication, shows how PacBio technology contributed to the findings, and outlines implications for future studies.