The Revio system offers faster and more cost-effective long-read sequencing for metagenomics researchers, allowing for more high-quality and complete microbial datasets and access to new frontiers of discovery.
The best in metagenomics just got better
Shotgun metagenomics with PacBio highly accurate long-read sequencing (aka HiFi sequencing) has been the premier approach for generating microbial datasets that are both very high quality and stunningly complete. Now, PacBio is changing the game all over again with the launch of the Revio system, a new long-read sequencing instrument that enables microbiome and metagenomics researchers to access exciting new avenues of discovery.
The Revio system takes the read length (up to ~25 kb) and read accuracy (99.9%) of the previous generation of PacBio sequencers and delivers it all at scale –faster and more cost effectively. At 15x the sequencing capacity with Revio, you can now access rich datasets flourishing with novel diversity and functional information that is simply waiting to be interpreted.
In this article, you will be introduced to the new ecosystem of solutions available on the Revio system that deliver a more seamless and powerful metagenomics experience, from sample to insight.
Topics covered include:
- New long-read specific solutions from Zymo Research for optimizing your sample preparation workflow to achieve better results.
- A fresh look at community profiling tools.
- Updated analysis pipelines for acquiring and evaluating more precise MAGs.
Long-read sample prep solutions offer a new level of certainty and reproducibility in microbiomics
A rigorous and well executed experimental design boosts confidence in results and enhances the influence and trust of the research community in the outcomes. You can now dial-in long-read sample preparation workflows at every step –with solutions from Zymo Research that ensure the inputs of a long-read sequencing study are of the highest quality with as little bias as possible.
- For sample collection and preservation, you can guard against community composition changes after sampling (even at ambient temperature) and preserve DNA fragment length with DNA/RNA Shield.
- At the DNA or RNA extraction step, you can confidently obtain quality extracts tailored to PacBio long-read sequencing lengths without introducing bias into the microbial profile with both manual and automated kits.
- To enable benchmarking and provide consistency across experiments, Zymo Research offers microbiome standards that can be used to detect biases in everything from extraction methods to library preparation and bioinformatic analysis.
Obtain more taxonomic and functional profiling information simultaneously
Because the Revio system offers a 15-fold throughput increase in long-read shotgun metagenomic information, the analysis phase of a HiFi microbiome study requires well-constructed software solutions to make the most of such rich data.
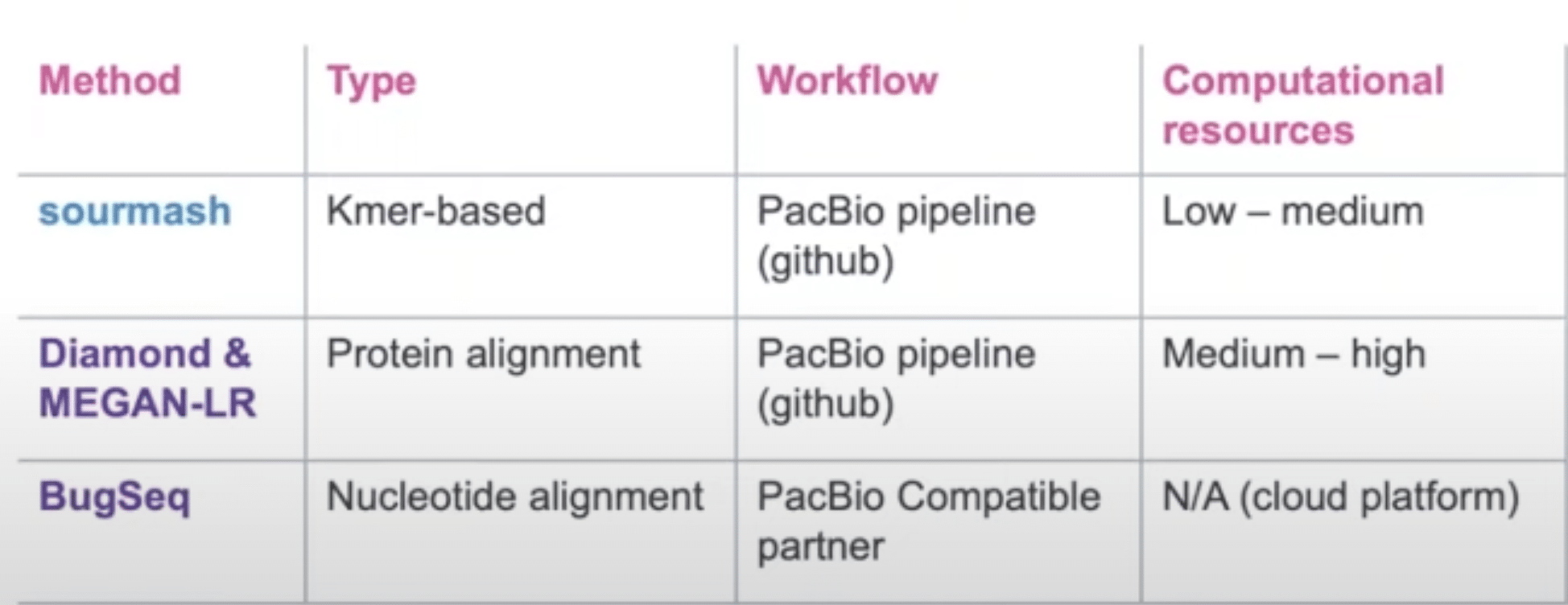
For community profiling, PacBio scientist Daniel Portik (@DPortik) and colleagues conducted a comparative analysis of numerous tools to determine which methods are the most adept at generating quality results from HiFi reads. In this study which compared precision and recall, Sourmash, Diamond + MEGAN-LR, and BugSeq were found to be the top performing. Sourmash offered the best tradeoff between precision and recall while Diamond + MEGAN-LR and BugSeq offered near-perfect precision with slightly reduced recall.
Because HiFi reads can span an average of eight genes at a time, the Diamond + MEGAN-LR pipeline enables you to assign taxonomic classification and functional annotations from the same sample, simultaneously.
Stemming from the outcome of this study, PacBio is pleased to offer workflow pipelines based on these three methods (see table below).
Capture more high-quality MAGs than ever
Most of our planet’s microbial diversity cannot be grown in a laboratory which makes metagenomic sequencing one of the only ways to study these unculturable organisms. Generating metagenome assembled genomes (MAGs) from a shotgun metagenome dataset is one of the best techniques for investigating these prokaryotic phantoms. For these assembly focused studies, PacBio long-read sequencing offers superior performance enabling researchers to generate more MAGs and more circular single-contig MAGs than short-read sequencing alternatives.
By using hifiasm-meta to create assembled contigs and the new completeness-aware HiFi-MAG-Pipeline v2.0, you can generate up to 186% more single contig MAGs than using just a single binning strategy like MetaBat2 and up to 28% more single contig MAGs than the previous v1.5 PacBio pipeline. The boost in quality and quantity of MAGs obtained from HiFi data make it possible to identify MAGs belonging to novel taxa as well as multiple MAGs from the same species which can indicate strain-level variation. This level of detail represents a significant leap forward for what can be achieved with a metagenome.
Boost MAG recovery at reduced costs with samples run on the new Revio instrument
The Revio system gives microbiome researchers more flexibility to do everything from multiplexing to deep sequencing of metagenomes at a cost-effective price point for labs great and small.
The next era of metagenomics starts now
With long-read tailored solutions that span the entire lifecycle of a metagenomics research program, from sample collection to data analysis, HiFi metagenomics is well on its way to becoming the method of choice for microbiome research. Improvements in sample prep optimization along with greater data yield and a more refined and flexible analysis experience give microbiologists access to a new world of possibilities that are waiting to be discovered.
Interested in learning more about metagenomics and long-read sequencing?
Obtain more species and MAGs from HiFi shotgun metagenomics data with updated pipelines
Cost-effective microbiome and metagenomic HiFi sequencing with PacBio service providers