
Full-length 16S and shotgun metagenome HiFi sequencing with certified PacBio service providers can deliver remarkably affordable microbiome and metagenomic data with explanatory power that is second to none.
Gaps, assembly errors, chimeras, and contamination are workaday challenges for microbiologists who need to produce references from metagenome sequencing data. With all the data filtering required one could imagine that there must be a better way. Luckily, HiFi sequencing solutions with PacBio service providers can help microbiome and metagenomics experts tackle these obstacles with premium data quality and surprising cost benefits.
Key takeaways
- Uncover the true diversity of complex microbiomes with stunningly accurate, species and strain-level detail
- Achieve high-precision taxonomy and rich functional information while recovering more high-quality metagenome assembled genomes (HQ MAGs) and more circular, single-contig MAGs than ever before –even at lower coverage with HiFi shotgun metagenome sequencing.
- Get started for as little as $20 USD/sample including PacBio amplicon library prep and sequencing
Species-level detail matters
PacBio full-length 16S sequencing on the Sequel IIe System delivers species-resolved information that would otherwise require shotgun approaches. Whether you study complex environmental samples or the human gut microbiome, get the clarity you need to connect community composition to variations in phenotype, ecosystem function, and more.
In a webinar earlier this year, George Weinstock (@geowei) of the Jackson Laboratory for Genomic Medicine discussed the importance of species resolution in microbiome research, highlighting how full-length HiFi 16S sequencing can resolve all the bacterial clades commonly found in the human gut microbiome, down to the species level.
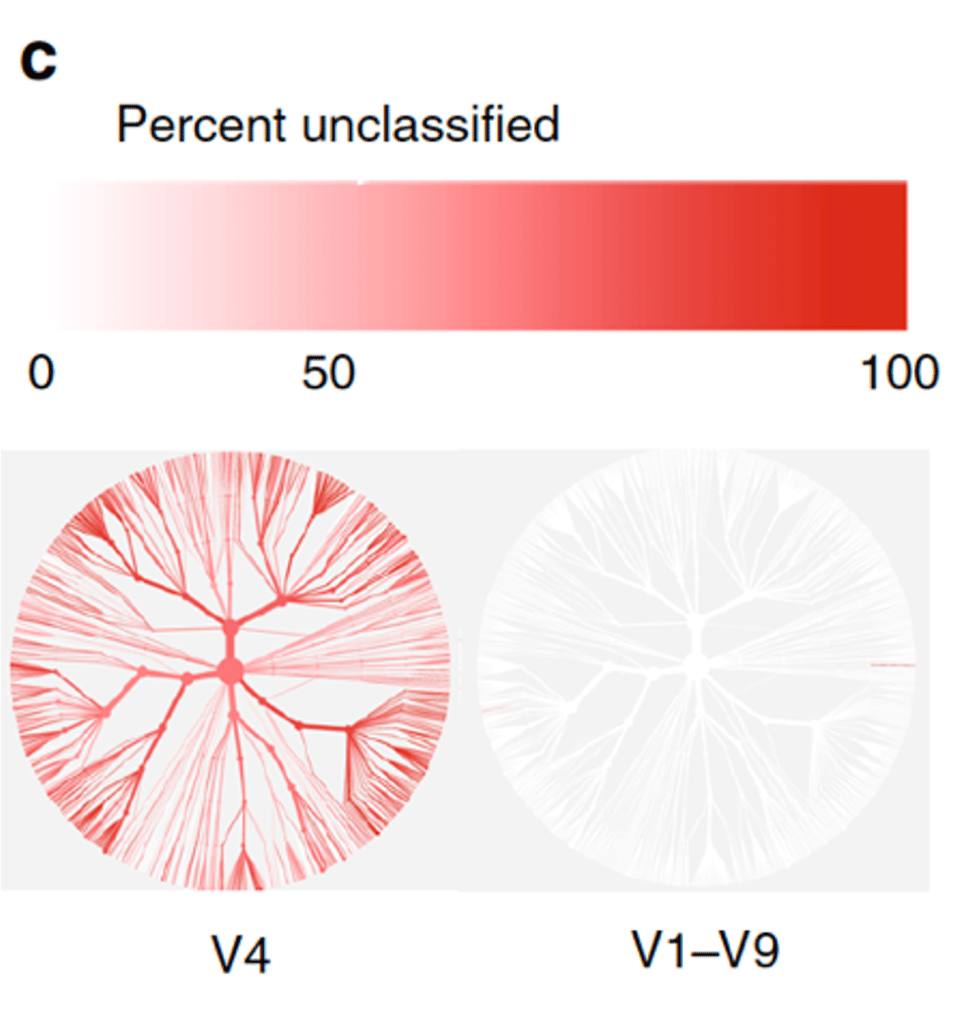
“With V1-V9, almost all [sequences] could be accurately identified to the species level. With V4, more than half could not be identified at the species level, so you are sort of locked into the genus level or higher… The full-length sequences are definitely the gold standard, there is no question about it.”
Using the 16S data from healthy stool donors as an example, Weinstock pointed out that even if a researcher can observe an accurate frequency of Bacteroides at the genus-level with a conventional sequencing approach, vital information will be missed due to the tremendous functional and ecological variation between microbial species (Figure 1) –and this is exactly what makes PacBio 16S HiFi reads so powerful. By sequencing around the same molecule multiple times, HiFi sequencing produces long, highly accurate, single molecule consensus reads for observing and understanding microbial communities down to the finest detail with complete 16S sequences.
Capture more information with less data
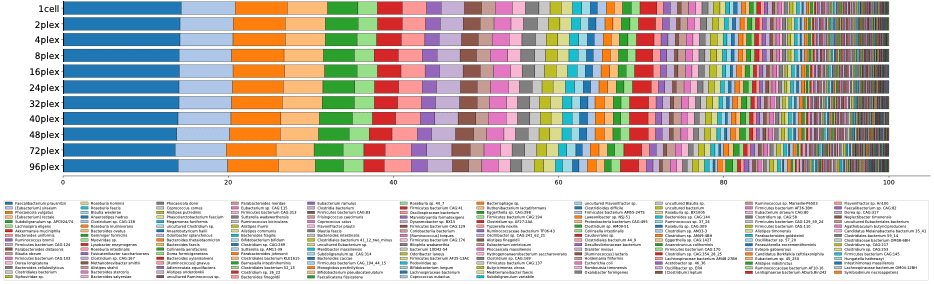
Downsampling experiments using PacBio’s newest human gut microbiome reference data set shows the number and relative abundance of bacterial taxa remains stunningly consistent down to as few as 60 K reads (0.5 Gbp), or 48 samples per SMRT Cell 8M.
How can HiFi sequencing be that efficient?
The high accuracy and long read lengths of HiFi sequencing deliver 6-8 complete genes per read, with nearly all adding actionable information (Figure 2) including functional assignments for approximately 90% of reads.
For assembly-focused projects, recent data from sequencing the ZymoBIOMICS TruMatrix fecal reference (D6323) shows that even modest amounts of HiFi sequencing data can have a significant impact on metagenome assembly quality and drive new insights, no matter what community you study. For as low as ~$10 USD† per high-quality (HQ) metagenome assembled genome (MAG), you can generate up to 35 HQ MAGs for up to 4 multiplexed human fecal samples or up to 110 HQ MAGs for 1 sample with only a single SMRT Cell 8M and many of which are single contig, complete, or near complete HQ MAGs.
Make the most of lab resources with HiFi sequencing
Avoid having to restart and reanalyze important projects and data. Get answers to challenging microbiology questions the first time –whether you are characterizing community composition, functional capacity, or assembling references for unculturable species. PacBio HiFi sequencing provides valuable insights that alternative approaches cannot deliver.
Get the benefits of HiFi 16S and metagenomics for your project
These PacBio certified sequencing service providers are just a few of many that can perform microbial genomics sequencing services with competitive pricing as low as $20 USD/sample for PacBio HiFi amplicon library prep and sequencing. Connect with a PacBio scientist for more information
Americas
• Dalhousie University Integrated Microbiome Resource (IMR)
• Yale Center for Genome Analysis (YCGA)
• Icahn School of Medicine at Mount Sinai Genomics Core Facility
• Arizona Genomics Institute
• BYU DNA Sequencing Center
• University of Louisville
• University of Maryland Institute for Genome Sciences
• University of Minnesota Genomics Center
• University of Illinois at Urbana-Champaign Biotechnology Center
• University of Delaware DNA Sequencing & Genotyping Center
• University of Wisconsin-Madison Biotechnology Center
• Mayo Clinic Genome Analysis Core
• Omics and Precision Agriculture Laboratory / Global Institute for Food Security
• University of Washington PacBio Sequencing Services
• University of Oregon Genomics and Cell Characterization Core Facility
Asia/Pacific
Australia
• The Australian Genome Research Facility (AGRF) and The University of Queensland (UQ)
• Genomics WA
China
• Biomarker Technologies
• Biozeron
• Majorbio
• Novogene
• MAGIGENE
Korea
• Macrogen
• DNA Link
• Theragen Bio
• NICEM, Seoul National University (National Instrumentation Center for Environmental Management, College of Agriculture and Life Sciences)
• CJ Bioscience
Taiwan
• Biotools
• TGIA
Europe, Middle East, and Africa
• EMBL
• Fasteris / Genesupport
• Fisabio (The Foundation for the Promotion of Health and Biomedical Research of Valencia Region, Spain)
• Functional Genomics Center Zurich (FGCZ)
• Lausanne Genomic Technologies Facility
• University of Bern NGS Platform
Explore these additional resources
- Check out our analysis pipelines for metagenomics and 16S that are publicly available on GitHub. These tools are constantly being improved, and user feedback is always welcome.
- Test drive HiFi shotgun metagenome datasets that are also publicly available. These include mock community reference standards as well as empirical samples.
- Listen to Dan Portik (@DPortik) talk about MAG (Metagenome Assembled Genome) recovery and metagenomic downsampling studies with HiFi sequencing in our recent Metagenomics Webinar on demand here.
- Take a look at our Metagenomics Solutions Brochure, Metagenome Sequencing Best Practices Guide, and easy-to-follow sequencing protocols and analysis recommendations.
†All prices are listed in USD and cost may vary by region. Pricing includes library and sequencing reagents run on Sequel II or IIe systems and does not include instrument amortization or other reagents.