The complete picture
Build a more comprehensive genomic view of microbial pathogens
The advent of Single Molecule, Real-Time (SMRT) sequencing has radically increased the understanding of microbial genomics, pathogenicity, and antimicrobial resistance for public health and surveillance. The PacBio systems deliver the most comprehensive genetic information available, and with SMRT sequencing, federal agencies, healthcare centers, and other public health organizations have the ability to:
- Generate complete genome assemblies and gain a deeper understanding of community-acquired and hospital associated infections and transmission.
- Use microbial epigenetics to further characterize global outbreaks, pandemics, and epidemics.
- Inform local and global preparedness and response to seasonal and annual infections by rapidly resolving viral populations.
- Characterize microbial communities with culture-free methods to support surveillance and tracking foodborne pathogens
Solutions Guide
A new standard in pathogen genomics
Detect, track, and characterize pathogens to a new standard of completeness and precision. PacBio solutions offer exceptional accuracy and long read lengths to support a wide range of pathogen surveillance applications.
Blog
THWART FOOD-BORNE CONTAMINANTS WITH THE UNMATCHED ACCURACY OF HIFI SEQUENCING
HiFi sequencing delivers the most accurate and comprehensive microbial sequence data possible to give food safety and public health professionals the clarity they need to better understand contaminants and make informed and timely decisions.
Blog
FASTER AND MORE AFFORDABLE SEQUENCING WITH THE HIFI MICROBIAL HIGH-THROUGHPUT WORKFLOW
Highly-accurate HiFi reads produced by PacBio long-read sequencers –such as the Sequel IIe and Revio systems– enable researchers to access entirely new levels of contiguity, completeness, and accuracy when assembling genomes.
Learn how other scientists have used SMRT Sequencing to advance public health:
- A living legacy of microbiology celebrates 100 years
- Keeping a close eye on MRSA: lessons learned from PacBio sequencing surveillance
- The antibiotic arms race: tracking K. pneumoniae in a hospital setting
- For pandemic surveillance, Labcorp scientists sequence thousands of SARS-CoV-2 samples with HiFi reads
- NARMS scientists track antibiotic resistance in foodborne bacteria using SMRT sequencing
- TB study finds some previously reported virulence variants were sequencing errors
- Scientists investigate minor influenza strains in pandemic, revealing new clues in flu transmission
Spotlight
NEW INSIGHTS INTO NDM RESISTANCE IN HEALTH CARE SETTINGS
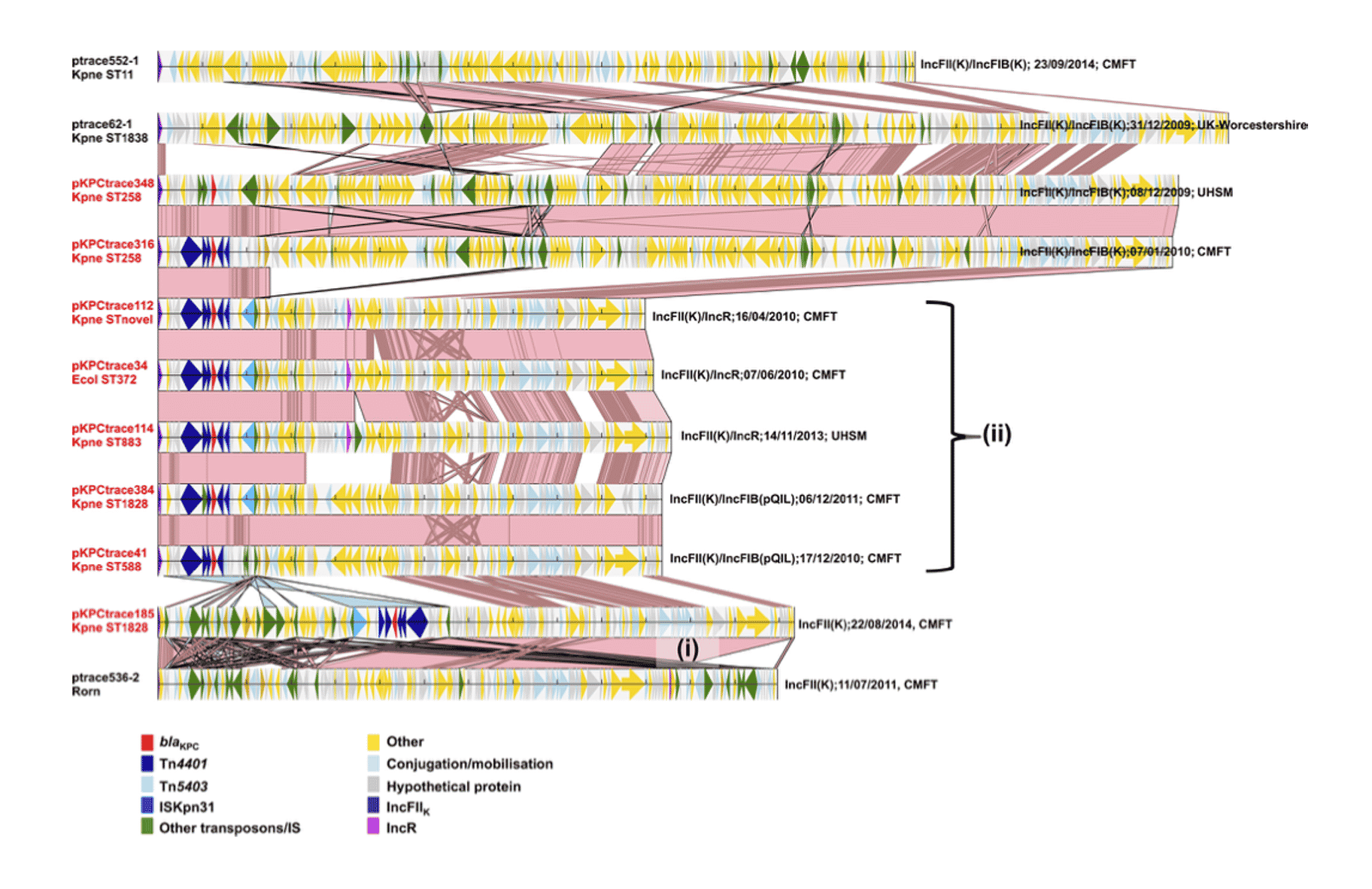
Large-scale sequencing and closed PacBio genomes link the spread and persistence of blaKPC carbapenem resistance in UK hospitals to the fluid exchange of mobile elements among plasmids capable of transfecting a broad range of hosts. The finding that horizontal transfer, rather than clonal expansion, is that main driver of increasing resistance has important implications for surveillance.
Stoesser, N., et. al. (2020) Genomic epidemiology of complex, multispecies, plasmid-borne blaKPC carbapenemase in Enterobacterales in the United Kingdom from 2009 to 2014. Antimicrob Agents Chemother, (64)5, p.e02244-19.
Spotlight
LONG READS SHED LIGHT ON THE SPREAD OF ANTIBIOTIC RESISTANCE IN FOOD SYSTEMS
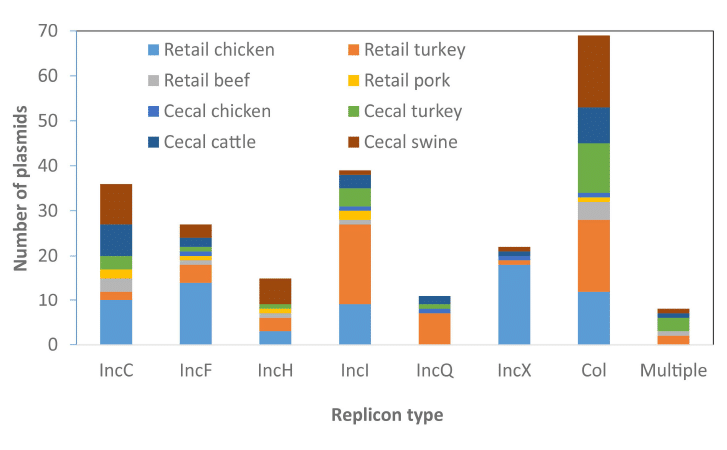
Long-read sequencing of 134 multidrug-resistant Salmonella strains revealed important trends and details about the spread of AMR resistance in our food systems, including chromosomal integration of plasmids and the potential for co-selection of AMR genes in the presence of heavy metals used in food animal production.
Li,C. et. al. (2021) Long-read sequencing reveals evolution and acquisition of antimicrobial resistance and virulence genes in Salmonella enterica. Front Microbiol, 12, doi: 10.3389/fmicb.2021.777817