The transcriptome you’ve been waiting for
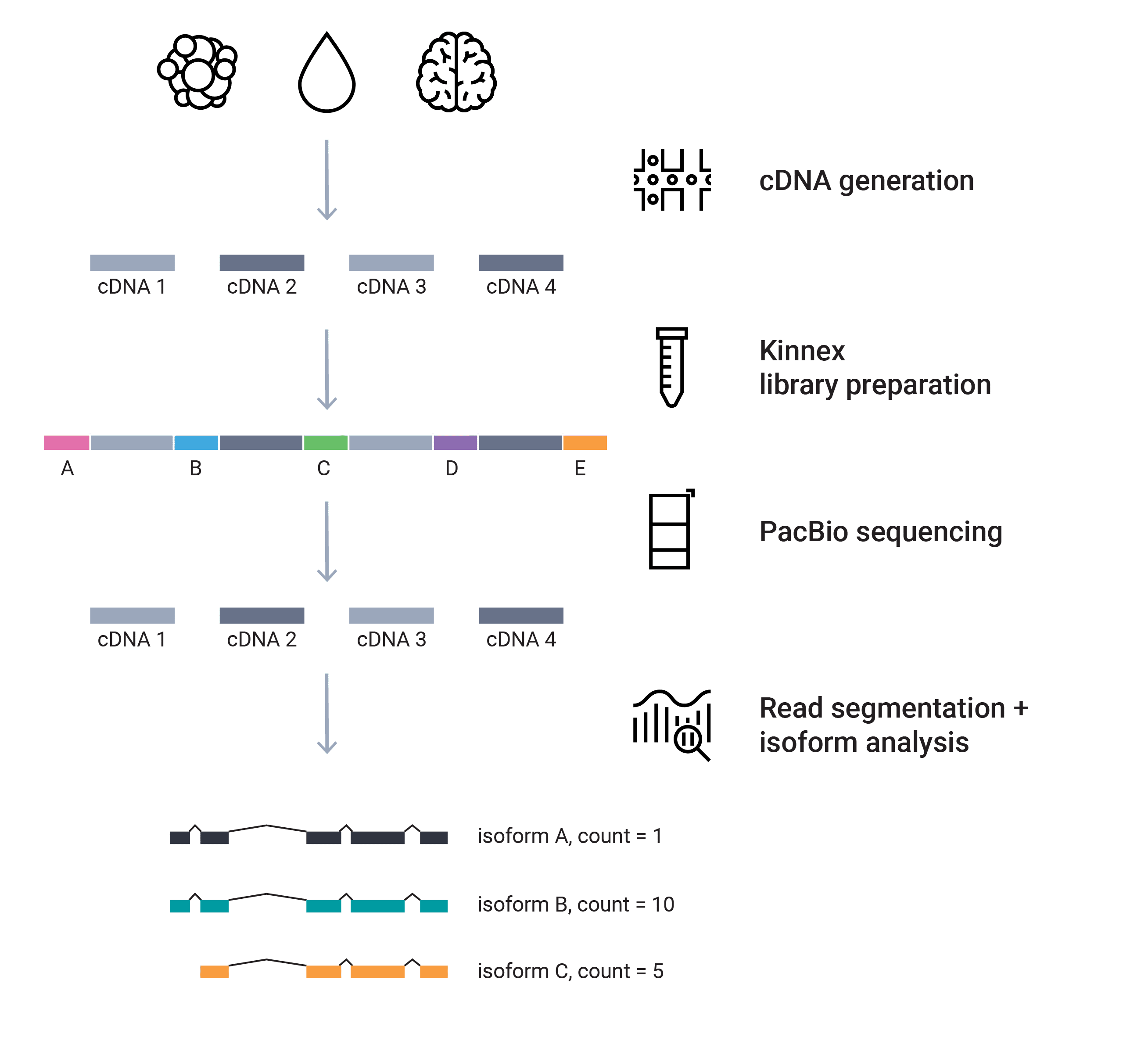
Full-length isoform sequencing for bulk and single-cell RNA studies allow for characterization of complex alternative splicing, fusion transcripts, prediction of open reading frames, and cell-type specific isoform expression. Short-read RNA-seq cannot sequence the entire transcripts and relies on computational methods for inferencing, often resulting in incomplete or missing transcripts that miss the true biology.
Other long-read technologies can be less accurate, producing fewer or more truncated reads (Pardo-Palacios et al. 2023, Calvo-Roitberg et al. 2023).1, 2 Get the most out of your RNA experiments with full-length RNA sequencing on PacBio.
Neurology
research
Fully characterize isoform diversity in neurons and glial cells and capture alternative splicing events and complex gene architecture in the brain. Understand mechanisms of brain development, neurological conditions, brain injury, and psychiatric disease with full-length RNA sequencing.
Rare disease research
Functionally characterize variants of unknown significance, discover aberrant splicing, differential transcript usage or expression patterns that reveal mechanisms of rare disease. Understand disease etiology or pathogenesis beyond germline DNA.
Cardiology research
Leverage a more complete cardiac transcriptome to fully characterize genetic factors underlying development of heart disease. Gain isoform-level resolution of cell types in the heart to better understand the mechanisms of cardiac function, injury, and disease.
Immunology research
Characterize complex splicing events and rearrangements in immune cells for a high-resolution profile of immune response. Profile antibodies and immune cell types, their microenvironment interactions, and response to treatment, disease, or pathogens.
APPLY NOW FOR THE HUMAN RNA SMRT GRANT
Ready to elevate your research with highly accurate full-length RNA information? Submit your application to win free sequencing.
Whitepaper
Bulk and single-cell isoform sequencing for human disease research
In this whitepaper, we review full-length RNA-sequencing with the PacBio Iso-Seq method and its applications in human genomics.
Human RNA sequencing applications
Novel mechanisms in neurology
Discover how customers are using full-length RNA sequencing with PacBio to power neurology research. This webinar includes sessions on neurobiology and cell atlas work, as well as translational research on neurodegenerative disease.
Driving discoveries in cardiology
Register for our webinar: Advancing cardiology research with long-read RNA sequencing. Discover how guest researchers are utilizing full-length RNA sequencing with PacBio to power groundbreaking research for a better understanding of heart development and cardiovascular injury and disease.

Understanding functional impact in Rare Disease
Full-length isoform sequencing for resolving the molecular basis of Charcot-Marie-Tooth 2A3
In this study, authors utilized full-length RNA sequencing with PacBio to elucidate the function of an intronic SNP thought to be causal in a case of axonal type 2a Charcot-Marie-Tooth disease. The study identified five distinct transcripts caused by alternative splicing, and found that while the canonical transcript was present, the candidate SNP led to deficient protein, a likely causal mechanism for the disease.
Uncovering insights in Immunology
Authors discuss limitations of other methods for adaptive immune receptor repertoire sequencing (AIRR-seq) and describe the use of PacBio sequencing to provide “highly accurate (mean read accuracy ~Q60) near full-length Ab sequences,” generating the most comprehensive view of bulk-expressed antibody repertoires to date.
Explore
Did you know we have a comprehensive library of articles, reports, papers, and videos?
- Calvo-Roitberg, E., Daniels, R. F., & Pai, A. A. (2023). Challenges in identifying mRNA transcript starts and ends from long-read sequencing data. bioRxiv, 2023-07. https://doi.org/10.1101/2023.07.26.550536.
- Pardo-Palacios, F. J., et al., (2023). Systematic assessment of long-read RNA-seq methods for transcript identification and quantification. bioRxiv, 2023-07. https://doi.org/10.1101/2023.07.25.550582.
- Stergachis, A. B., et al. (2023). Full-length isoform sequencing for resolving the molecular basis of Charcot-Marie-Tooth 2A. Neurology Genetics, 9(5). https://doi.org/10.1212/NXG.0000000000200090
Blog
Into the multiome
Leverage full-length RNA sequencing in multiomics for an extraordinary view of the human genome. Leveraging other PacBio workflows, discover how you can simultaneously capture the human genome, transcriptome, CpG methylome, and chromatin epigenome, all on a single Revio SMRT Cell sequencing run.
FULL-LENGTH RNA SEQUENCING SPOTLIGHT
KINNEX FULL-LENGTH RNA KIT ENABLES SCALABLE, COST-EFFECTIVE ISOFORM SEQUENCING
The Kinnex full-length RNA kit produces full-length isoforms up to 10 kb. Combine with the Iso-Seq express 2.0 kit and SMRT Link analysis to characterize and compare known and novel isoforms for your RNA research.