As Marvel superheroes traverse the multiverse to save the day, genomics researchers are our superheroes as they navigate the daunting multiverse that is biology. The complex and dynamic interactions between the genome sequence, its epigenetic regulation, and their combined effects on transcript expression and splicing are fundamental to our understanding of biology, and their aberrations to elicit disease. Multiomic approaches allow us to better understand the underlying connections and flow of information from the genome to the transcriptome. While now commonplace in biomedical research, they have been hampered by requiring multiple assays, sequencing runs, technologies, and combining separate datasets, making such studies costly and labor-intensive.
Today, we are excited to share a new superpower for multiomic research. From a collaboration led by Dr. Andrew Stergachis and Dr. Mitchell Vollger at the University of Washington, a new preprint, entitled Synchronized long-read genome, methylome, epigenome, and transcriptome for resolving a Mendelian condition, describes the simultaneous generation of four high-quality, haplotype-resolved ‘omes – the genome, CpG methylome, chromatin epigenome, and transcriptome – from a single Revio SMRT Cell sequencing run.
This new capability is achieved by combining three advances. First, we leverage the higher throughput of the PacBio Revio system, yielding high-quality long-read HiFi sequence data at scale. Second, the UW researchers developed a machine learning approach for chromatin Fiber-seq data (Stergachis et al. 2020) to accurately assemble haplotype-resolved, single-molecule chromatin accessibility, nucleosome positioning, and transcription factor occupancy patterns. Third, we adapted a library protocol from the MAS-Seq method (Al’Khafaji et al. 2023) to concatenate full-length cDNA molecules to increase the throughput for full-length RNA sequencing. The concatenated cDNA library targeted a similar fragment size to standard HiFi WGS libraries, which allowed combining the Fiber-Seq treated gDNA library with the matching cDNA library in a single HiFi sequencing run on the Revio system.
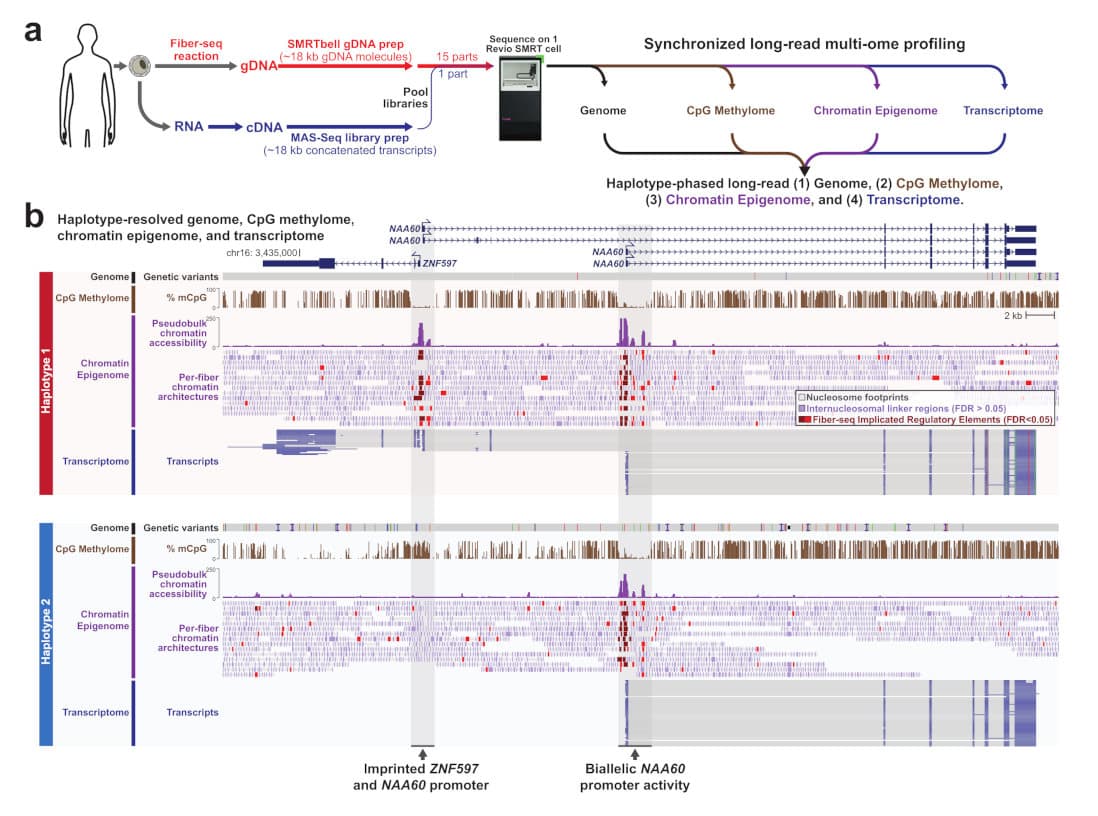
We first benchmarked the performance of this synchronized multiome assay against Genome-in-a-Bottle (GIAB) samples with known ground truths, demonstrating that the Fiber-seq data retain highly accurate calling of all variant types (SNVs, Indels, and SVs), CpG methylation detection, and diploid de novo genome assembly quality (haplotype contig N50s >20 Mb, consensus accuracy QV 50). Deconcatenated cDNA reads sequenced in the same Revio SMRT Cell provided matching full-length RNA transcript data for these GIAB samples.
We next applied this multiomic approach to cells from a participant within the Undiagnosed Diseases Network (UDN) with unexplained developmental delay, polymicrogyria, sensorineural hearing loss, and bilateral retinoblastomas, harboring a known chromosome X-13 balanced translocation of uncertain significance. The multiome data from a single Revio SMRT Cell showed that each ‘ome contributed to the explanation of the molecular basis for the multiple disease symptoms of the individual, and revealing the involvement of four different genes in the multitude of phenotypes.
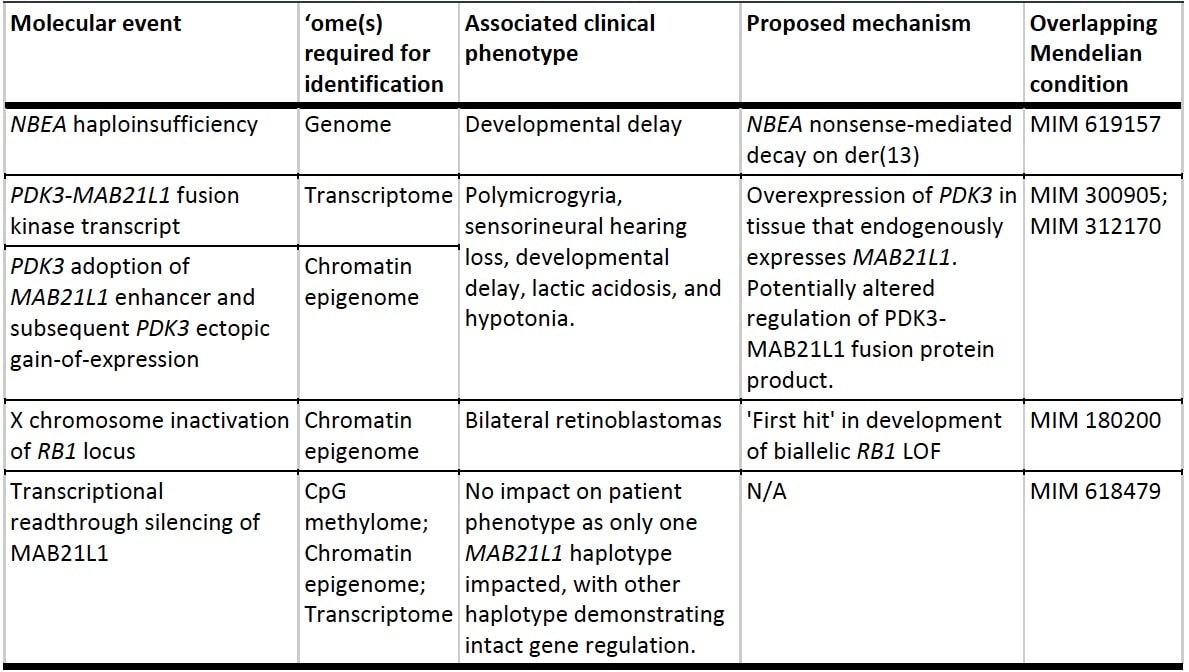
These results demonstrate the strong utility of an integrated, single-technology long-read multiomic profiling for resolving the mechanisms underlying unexplained rare conditions. While demonstrated here for a human genome with a previously unexplained disease, this new synchronized multiome paradigm can be applied to any biological sample for which a deeper understanding of its molecular fabric is sought after. So, we see great opportunities for this paradigm for areas such as cancer, plant and animal genomes, population genomics, and many others. We are very excited to see what discoveries and understanding await in all these areas by simultaneously elucidating so many dimensions of a biological sample – getting to know everything, everywhere, all at once.
Connect with us at ASHG
Please see us at the upcoming ASHG conference, which will include a presentation by Dr. Stergachis at our workshop, and more news about product offerings for sequencing full-length RNA with scalable throughput. Please connect with us as you plan to dive into the multiverse of genomics –we are here to help you in your superhero mission!