More samples, one SMRT Cell
HiFi sequencing is the ideal long-read technology for multiplex sequencing. The high base accuracy allows confident identification of indexes, plus the on-instrument demultiplexing option simplifies and streamlines downstream data analysis. All that is required to multiplex is the inclusion of a short index (a barcode, typically 10 bp in length) adjacent to the SMRTbell adapter (Figure 1). This index can be added to each sample during the DNA library construction process using one of the 384 SMRTbell adapter indexes (Figures 1-3), upstream during sample prep using a PCR primer (Figure 5, 7), or with an indexed adapter from a compatible partner (Figure 4).
Whole genome sequencing
The increased throughput of the Revio system offers increased multiplexing capacity and corresponding reductions in per sample sequencing costs.
Kinnex
The Kinnex kits provide scalable, cost-effective RNA sequencing for bulk RNA, single-cell RNA, and 16S rRNA sequencing.
Targeted sequencing
Targeted long-read sequencing enables researchers to interrogate the most challenging genomic regions at scale.
Multiplexing for whole genome sequencing
The increased throughput of the Revio system offers increased multiplexing capacity and corresponding reductions in per sample sequencing costs. Whether for variant detection of the human genome or assembling hundreds of microbial isolates, the SMRTbell adapter index plates 96(A, B, C, and D) provide an easy and streamlined solution to sample barcoding to enable multiplexing with no additional prep step. Simply use the index adapter in the ligation step, which is included in the HiFi prep and HiFi plex prep kits (Figure 1).
Figure 1: The SMRTbell hairpin and 10 bp index sequence flanking a DNA insert.
Kinnex
The Kinnex kits provide scalable, cost-effective RNA sequencing for bulk RNA, single-cell RNA, and 16S rRNA sequencing. Kinnex kits utilize the MAS-Seq method, where small amplicons are concatenated in an ordered array into larger fragments to fully utilize HiFi long read lengths and increase throughput. There are three Kinnex kits that utilize this concatenation method: Kinnex full-length RNA kit, Kinnex 16S rRNA kit, and Kinnex single-cell RNA kit. Visit https://www.pacb.com/technology/kinnex/ to learn more.
All three kits utilize the same set of 4 barcoded Kinnex adapters to enable library-level multiplexing. Each application enables amplicon-level multiplexing. The Iso-Seq express 2.0 kit enables 12-plex barcoded cDNA, while the 16S amplicon protocol enables 384 unique samples.
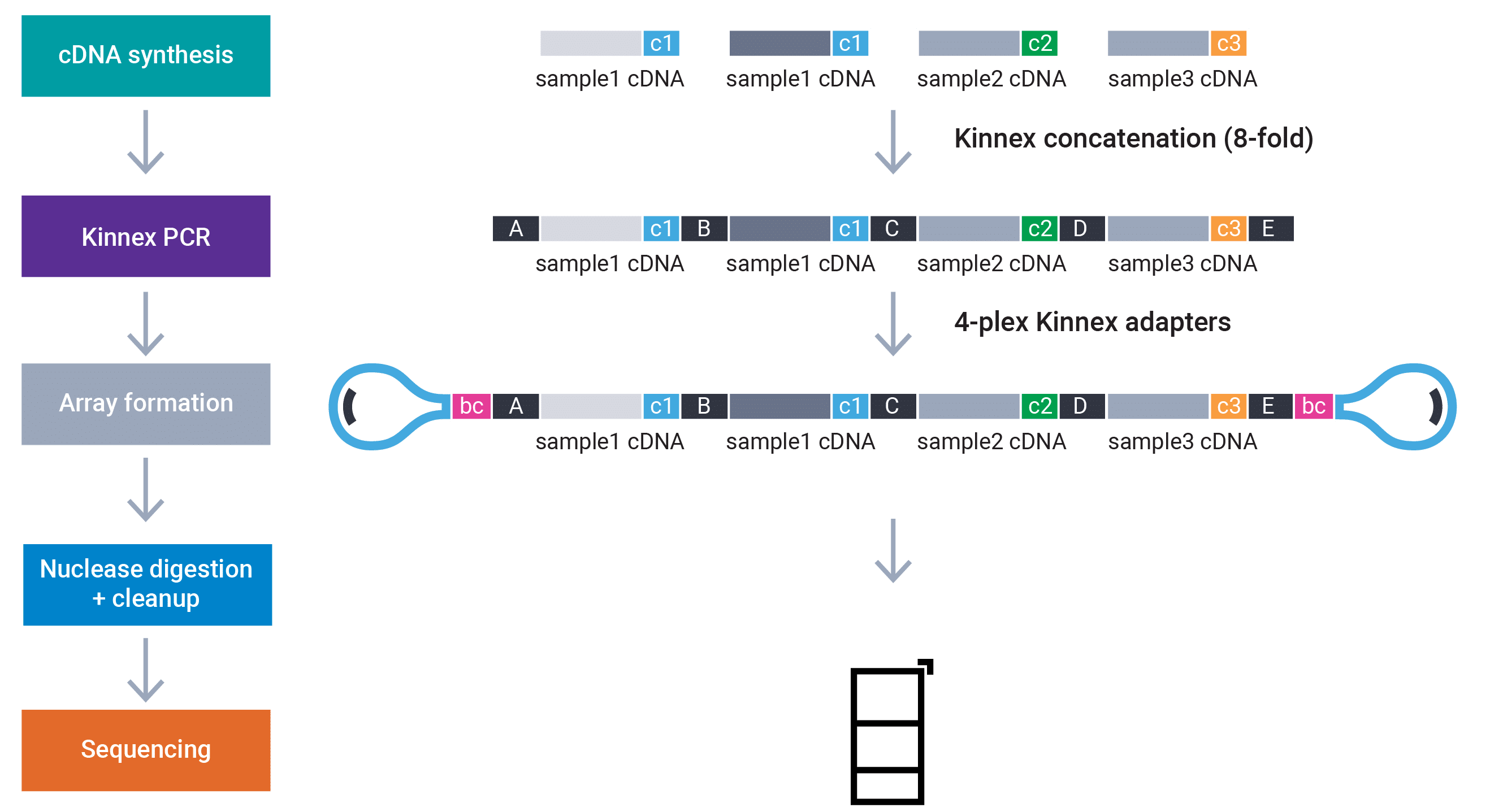
Figure 2: The Kinnex barcoded adapters are shown in pink (bc) at the two ends of the double-stranded structure. Note that for full-length RNA sequencing, additional multiplexing is enabled through the use of barcoded cDNA primers (denoted c1, c2, c3) in the figure. The segments A, B, … E are illustrations of the Kinnex concatenation primers, and, while used to create the Kinnex arrays, cannot be considered as means for multiplexing.
Targeted sequencing
Targeted long-read sequencing enables you to interrogate the most challenging genomic regions at scale. PacBio offers different targeting methods to meet your application needs. PureTarget panels enable PCR-free targeting using CRISPR/Cas9 method (Figure 3). With our partner, Twist, we offer HiFi target enrichment using probe-based hybrid capture (Figure 4). PCR amplicons are low cost, high throughput and offer diverse barcoding strategies (Figure 5 – 8). Learn more about targeted HiFi sequencing.
PureTarget libraries
PCR-free target enrichment with CRISPR-Cas9 system is available with the PureTarget repeat expansion panel. These libraries retain methylation at CpG sites, have no GC bias and less size bias compared to other targeting methods that use PCR. Sample multiplexing uses SMRTbell adapter index plates.
Learn more in our App note
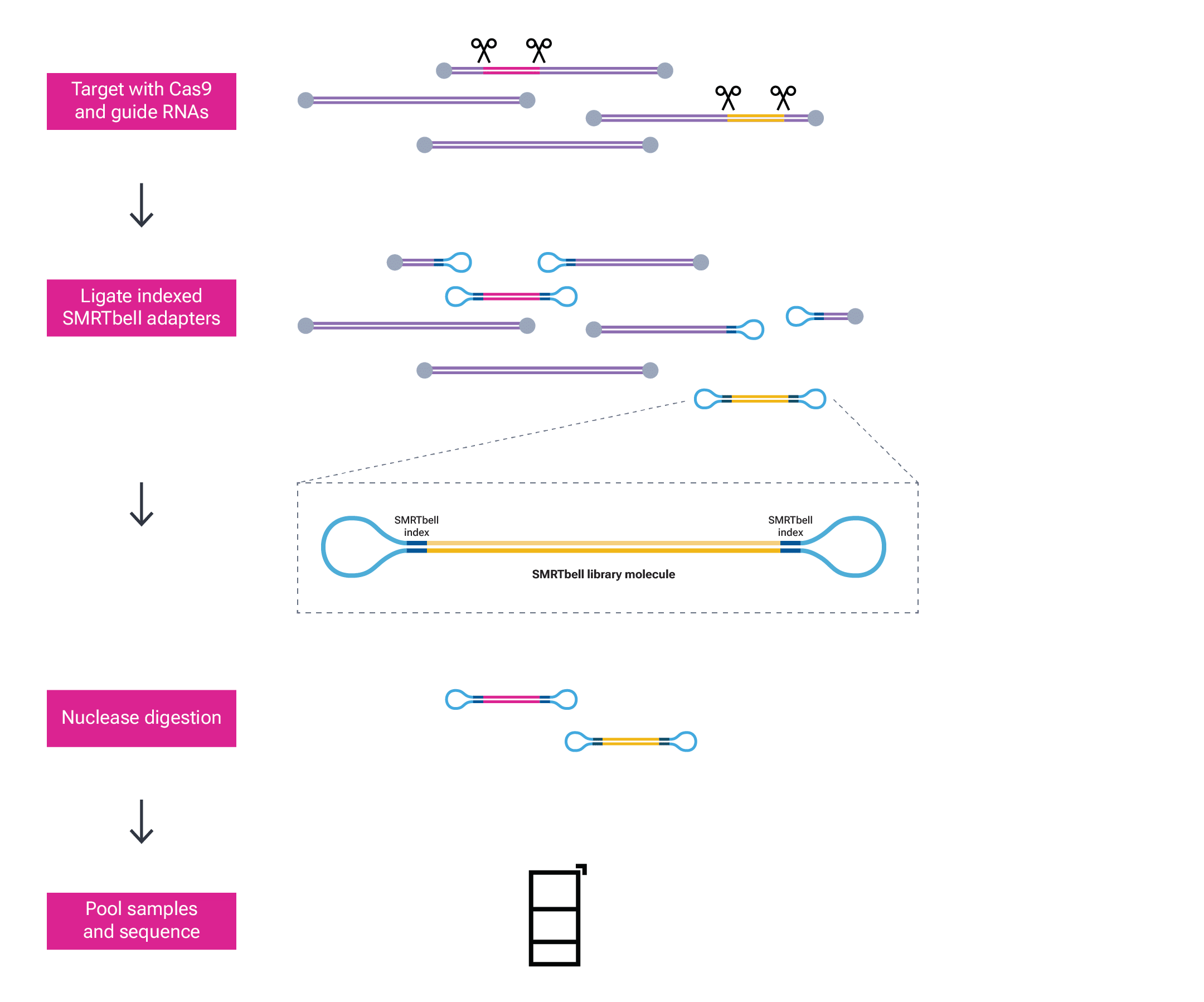
Figure 3: Targeting is done using Cas9 and a pair of guide RNAs flanking the region of interest. Samples are barcoded by ligating indexed SMRTbell adapters during library prep. Barcodes are shown in dark blue. Nuclease treatment removes non-SMRTbell templates prior to sample pooling and sequencing.
HiFi Target Enrichment with Twist Bioscience
PacBio offers HiFi target enrichment using probe-based hybrid capture from our partners at Twist Bioscience (Figure 4). This approach enables high throughput and comprehensive variant calling including structural variants and phasing. Learn more in our App note, and Twist protocol.
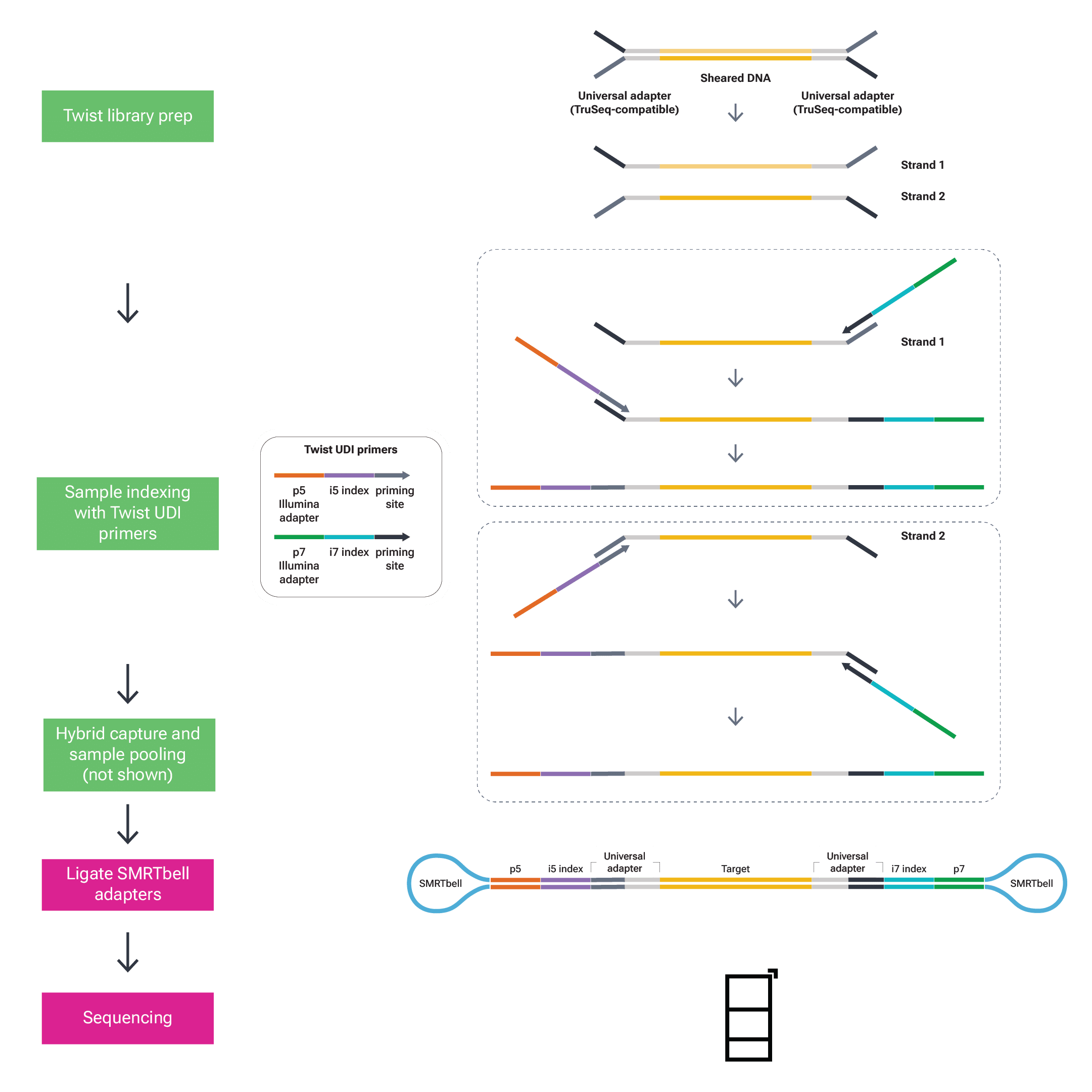
Figure 4: During Twist library prep, universal adapters are ligated to sheared DNA and P5 and P7 adapters (orange and green) with i5 and i7 indices (purple and cyan) added with PCR. After hybrid capture to target regions of interest (not shown) samples are pooled, SMRTbell adapters are ligated in PacBio HiFi library prep, and samples are sequenced.
PCR amplicons with indexed primers
A streamlined and cost-effective targeted sequencing approach is PCR with barcoded primers. Learn more in our App note and protocol. Recommended barcode sequences to support 384 dual-indexed samples available.
![]()
Figure 5: Target-specific primers with barcode tails (purple and teal) are used to amplify and index a region of interest. Samples are pooled and a single PacBio library is prepared for sequencing.
PCR amplicons with indexed adapters
A streamlined targeted sequencing approach for existing PCR assays is to use indexed SMRTbell adapters. With the automated HiFi plex prep kit, prepare individual SMRTbell libraries for each sample, pool, and sequence on one SMRT Cell. Learn more in our PacBio protocol.
![]()
Figure 6: Target-specific primers are used to amplify a region of interest. Barcode sequences (dark blue) are added during HiFi library prep.
PCR amplicons with M13 indexed primers
A flexible and cost-effective targeted sequencing approach is to add indices with PCR and barcoded M13 primers. Learn more in our App note. Additional resources: PacBio protocol, barcode fasta file for demultiplexing and M13 primer plate layout.
![]()
Figure 7: Target-specific primers with M13 tails are used to amplify a region of interest. Dual indices (pink and teal) are added with barcoded M13 primers available from PacBio in a pre-mixed plate. Samples are pooled and a single PacBio library is prepared for sequencing.
Indexed primers and adapters
If more than 384 samples per SMRT Cell are required, A combination of indexed primers and indexed SMRTbell adapters may be used. Uniquely indexed amplicons are pooled in a PacBio HiFi library prepared with indexed SMRTbell adapters. Using the same set of indexed primers for each indexed SMRTbell adapter, HiFi libraries may then be pooled and sequenced on one SMRT Cell. Two round of demultiplexing is required: round 1 demultiplexes SMRTbell adapter indices, and round 2 demultiplexes primer indices.

Figure 8: Sample indices are added in two steps of the workflow. First barcoded primers are used, (purple and teal), then indexed SMRTbell adapters are added (dark blue).
