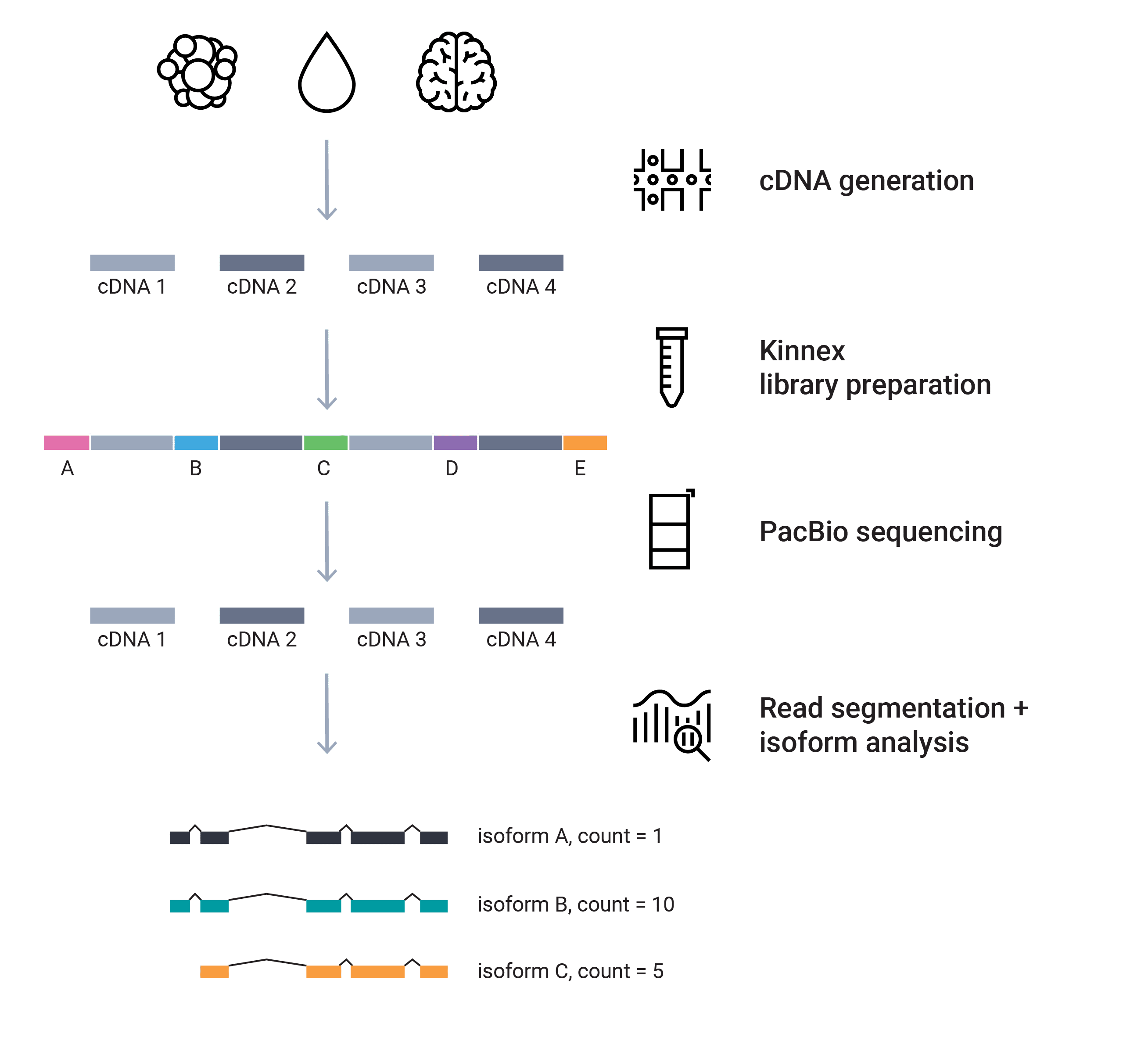
RNA sequencing
The future of RNA sequencing is with long reads! The Iso-Seq method sequences the entire cDNA molecules – up to 10 kb or more – without the need for bioinformatics transcript assembly, so you can characterize novel genes and isoforms in bulk and single-cell transcriptomes and further:
- Characterize alternative splicing (AS) events, including alternative start sites, end sites, intron retention, and exon-skipping events
- Find gene fusions in tumor samples
- Identify allele-specific isoforms
- Detect differentially expressed isoforms and isoform switching events
- Predict functional impact of novel isoforms through open reading frame (ORF) prediction
On-demand webinar
UNDERSTANDING CLONAL EVOLUTION USING GAME THEORY AND SINGLE-CELL LONG-READ ISOFORM ANALYSIS
See how cancer researchers use the MAS-Seq for 10x Single Cell 3’ kit (now the Kinnex single-cell RNA kit) to identify driver mutations and trace clonal populations over time in a patient’s research sample who progressed from MDS (Myelodysplastic syndrome) to AML (acute myeloid leukemia).
Human
transcriptome studies
Explore how changes in isoform usages contribute to phenotypic differences between health and disease.
Plant + animal
transcriptome studies
Advance breeding programs and basic research with full-length cDNA sequencing that does not require a reference genome.
Single-cell
transcriptome studies
Visualize biology at an even higher resolution by capturing a more comprehensive and complete transcript sequence from individual cells.
“Long-read sequencing results in more accurate measurements than any obtained from short-read RNA-seq to determine the presence or absence of specific isoforms.”
Schertzer et al., Cas13d-mediated isoform-specific RNA knockdown with a unified computational and experimental toolbox, bioRxiv (2023)
The right solution for your RNA applications
RNA sequencing datasets
| Application | Dataset | Download literature | Technology | Sequencing system |
|---|---|---|---|---|
| Kinnex single-cell | Homo sapiens - PBMC 10x Chromium Single Cell (5' and 3' libraries) | Kinnex single-cell application note | HiFi long-read | Sequel II and Revio systems |
| Kinnex single-cell | Homo sapiens - HG002 (10x 5') | Kinnex single-cell RNA application note | HiFi long-read | Revio system |
| Kinnex full-length RNA | Homo sapiens – UHRR | Kinnex full-length RNA application note | HiFi long-read | Sequel II and Revio systems |
| Kinnex full-length RNA | Homo sapiens – HG002 | Kinnex full-length RNA application note | HiFi long-read | Revio system |
| Kinnex full-length RNA | Homo sapiens - Heart | Kinnex full-length RNA application note | HiFi long-read | Revio system |
| Kinnex full-length RNA | Homo sapiens - Cerebellum | Kinnex full-length RNA application note | HiFi long-read | Revio system |
Long-read RNA sequencing in action
Article
ISO-SEQ METHOD DIFFERENTIATES PSEUDOGENES ASSOCIATED WITH DISEASES
Read how researchers at University College London used the Iso-Seq method to accurately map and characterize the parent-pseudogene pair GBA-GBAP1 which is implicated in Gaucher’s disease and Parkinson’s disease.

Isoform-resolution single-cell atlas of colorectal cancer
Researchers apply long reads to single-cell RNA-Seq in colorectal cancer to identify recurring neoepitopes as potential cancer vaccine candidates.

ISO-SEQ METHOD OUTPERFORMS OTHER LONG-READ METHODS IN BENCHMARKETING CONSORTIUM STUDY
The LRGASP consortium published their assessment of long-read RNA sequencing using different sequencing platforms and analysis tools.

PROTEIN ISOFORM DISCOVERY ENHANCED BY ISOFORM SEQUENCING
Researchers use Iso-Seq data to predict open reading frames integrated into a proteogenomics workflow to discover novel protein isoforms to reveal the functional impacts of alternative splicing.
EXPLORE
Did you know we have a comprehensive library of articles, reports, papers, and videos related to RNA sequencing with the Iso-Seq method?
FULL-LENGTH RNA SEQUENCING SPOTLIGHT
KINNEX FULL-LENGTH RNA KIT ENABLES SCALABLE, COST-EFFECTIVE ISOFORM SEQUENCING
The Kinnex full-length RNA kit produces full-length isoforms up to 10 kb. Combine with the Iso-Seq express 2.0 kit and SMRT Link analysis to characterize and compare known and novel isoforms for your RNA research.
COMMON QUESTIONS ABOUT PACBIO ISO-SEQ METHOD AND KINNEX KITS FOR RNA SEQUENCING
Shaping the future
A HiFi movement: Isoform-resolved cell atlases
A shift to higher-quality, isoform-resolved cell atlases with the increasing adoption of full-length RNA sequencing with PacBio.
FEATURED LONG-READ SEQUENCING SYSTEMS
Achieve highly accurate full-length cDNA sequencing with PacBio long-read sequencers.