 Pseudogenes are typically regarded as non-functional DNA elements, and have therefore not received as much attention as their parent genes. Thanks to PacBio HiFi sequencing, many pseudogenes now RISE FROM THE DEAD by having their biology more fully elucidated.
Pseudogenes are typically regarded as non-functional DNA elements, and have therefore not received as much attention as their parent genes. Thanks to PacBio HiFi sequencing, many pseudogenes now RISE FROM THE DEAD by having their biology more fully elucidated.
As a recent example, in a new preprint entitled Pseudogenes limit the identification of novel common transcripts generated by their parent genes, researchers from University College London and eighteen collaborating institutions investigate the impact of pseudogenes on transcriptome analyses, using the parent-pseudogene pair GBA-GBAP1 (biallelic mutations in GBA1 result in Gaucher disease, and heterozygous mutations are among the most important genetic risk factors for Parkinson’s disease (PD) and PD progression). They observe only 42% of short reads mapping uniquely to GBA1, with the remaining reads mapping primarily to GBAP1 (the pseudogene). They observed that “this resulted in a significant misestimation of the relative expression of GBA1 to GBAP1.”
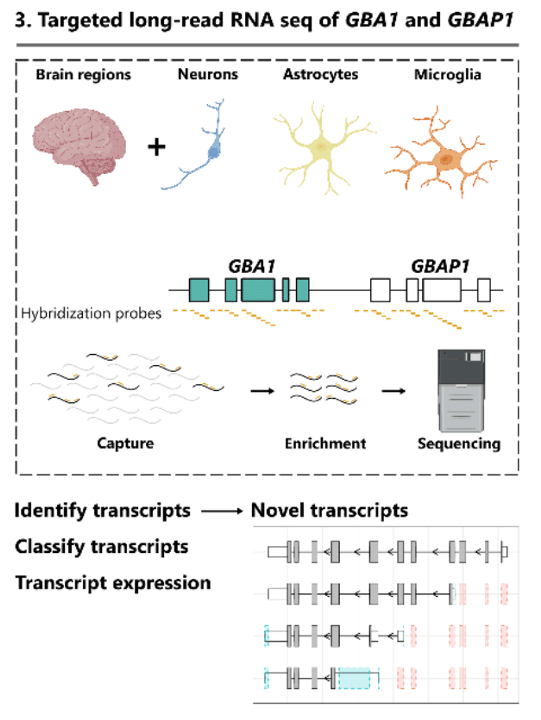
To obtain clearer insights into GBA-GBAP1 transcription, “PacBio Iso-Seq was used due to the high base pair accuracy (>99% accuracy) enabled by circular consensus sequencing (CCS) reads, which in turn, allows accurate mapping.” Using targeted Iso-Seq of 12 human brain regions, the researchers “identified 18 GBA1 transcripts that had a novel open reading frame (ORF) and 7 GBAP1 transcripts predicted to encode a protein, despite GBAP1 being classified as a pseudogene”, and including significant differences in GBAP1 ORF usage across different brain cell types.
Finding that inaccuracies in annotation from short-read sequencing are common for parent genes on a genome-wide scale, the authors conclude: “Given that 734 (17%) genes causing Mendelian disease have at least one pseudogene, these findings significantly impact our understanding of human disease and highlight the need for long-read RNA sequencing analyses at many loci.”
The work follows earlier studies that highlight the wide-spread occurrence of pseudogenes being active and potentially having important functions related to health vs. disease. E.g., for another FA-BOO-LOUS paper see Long-read cDNA sequencing identifies functional pseudogenes in the human transcriptome, which used the Iso-Seq method to identify hundreds of novel transcribed pseudogenes expressed in tissue-specific patterns in normal human tissues and cancer cell lines. These include pseudogene transcripts that have intact open reading frames and are translated in cultured cells, representing unannotated protein-coding genes. The authors thereby highlight “pseudogenes as a complex and dynamic component of the human transcriptional landscape.”
As we see RNA driving research for novel therapeutic approaches, it is more important than ever to have a comprehensive transcript map. HiFi reads are no TRICK, so we invite you to TREAT your research with long and accurate HiFi sequencing to resolve even the most challenging GHOSTS that lurk in the genome. And with our new MAS-Seq solution, single-cell resolved full-length transcript isoform expression studies are now more MAGICAL than ever before.
Interested in learning more?
Register to hear more about Emil Gustavsson, PhD, Postdoctoral Research Fellow’s work at the University College London here.