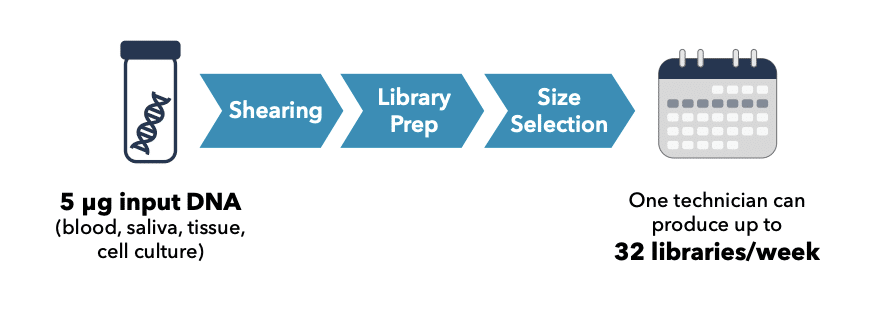
VAMOS! Discover hidden VNTRs with long reads
See how the Chaisson lab created a new method to characterize human VNTR variation with PacBio long-read sequencing. Structural variants are hard to find with short-read sequencing methods At present,…