
They spoke about omentum, chemosynthesis, chromothripsis, and… Tasmanian devils? This year’s virtual two-day SMRT Leiden Scientific Symposium and Informatics Developers Meeting was certainly educational.
With the pandemic and increased difficulty in being able to connect in person, we wanted to provide a forum for young investigators, post docs, and faculty to come together and share their research experiences during these abnormal times. The result? 27 speakers—the majority of whom were young investigators—sharing data and discoveries, and their advice for early-career scientists.
There was a great spectrum of presentations. The first keynote featured fun facts about Dominette (first Bos taurus to have her genome sequenced) and efforts to create a cow pangenome. Hubert Pausch of ETH Zürich discussed the downfalls of reference-guided variant discovery and the benefits of genome graphs to overcome some of the biases of linear mapping.
The second keynote by 2019 Human Genetics SMRT Grant winner Tychele Turner (@tycheleturner) highlighted the value of HiFi reads in investigating neurodevelopmental disorders, including 9p minus disorder and autism. Turner said HiFi reads should become the new paradigm because of their ability to detect more variants, reveal novel variation, and phase even the most complex genes.
“PacBio long-read sequencing is ushering in a new era in human genomics.” — Tychele Turner, PhD., Washington University Genetics
In other sessions, early data from another SMRT Grant winning project about neurological diseases with complex structural rearrangements was reviewed. Matthew Hestand of Cincinnati Children’s Hospital Medical Center was joined by University of Louisville researcher Corey Watson (@ctwatson29), who spoke about characterizing immunoglobulin haplotype diversity and its influence on the antibody repertoire, and Gloria Sheynkman (@GSheynkman) of the University of Virginia, discussed the integration of long-read RNA-Seq and mass spectrometry.
In another session, Harald Gruber-Vodicka (@GruberVodicka) of the Max Planck Institute for Marine Microbiology imparted some useful advice about sample preparation and assembly of the tiniest samples, and Jannat Ijaz (@sciencejannat) of the Wellcome Sanger Institute described catastrophic genome fragmenting events that can occur in cancer, and how she pieced together 900 fragments from esophageal organoids.
Lastly, in view of the pandemic and the challenges associated with it, we invited Melissa Smith (@SmithLab_UofL) to speak about work in her new lab at the University of Louisville, covering both COVID-19 surveillance and research into one of the biggest hurdles in HIV therapy, HIV “reservoirs.”
Pursue Your Passions
In addition to getting a look at the research being done around the world, we understand that the pandemic has been an unprecedented time for early-stage researchers. In view of these challenges, we decided to host a session dedicated to career advice, guidance, and open discussion. This ended up being one of the event’s most popular sessions, featuring valuable learnings from speakers.
Here are a few examples of the advice they shared:
On the art and craft of being a scientist, finding a mentor, and how to lead:
Choose your mentor over your scientific subject. Learning the craft and being the best scientist you can be is most important, and can then be applied to the subject you love. Also, don’t stymy your creativity. Practice it. Try to have one new idea every day—it doesn’t even matter if it involves science. — Jonas Korlach, PacBio CSO
Think about what kind of leader you want to be. You learn how to be a good scientist or bioinformatician, but no one tells you how to lead a group. Consider taking a course, or learn from your favorite leaders. — Susan Kloet, Leiden University Medical Center
On recognizing the talents we each possess, and using those talents to succeed in science:
Drive yourself. Don’t compare yourself to anyone else. Don’t rely on anyone else to set expectations and hold yourself accountable. Remove the word ‘should’ from your career vocabulary. — Melissa Smith
Be open to trying something new. And don’t be cowed by others’ successes. They’ve encountered challenges too. — Tychele Turner, PhD., Washington University Genetics
On being open to revision and excited by the chance to iterate:
Be happy if you see your papers returned covered in red (edits) – it means your supervisor cares. Don’t be frustrated. Take their advice and use it to improve. — Hubert Pausch, Prof. Dr., ETH Zurich
And, last but not least – on passion:
You can still make great contributions to science outside of academia. Even extracurricular activities like roller derby can teach you key soft skills like leadership and organization. Pursue all your passions. — Sarah Kingan, Senior Product Manager at PacBio
And, don’t forget to leave the lab every once in a while.
Bioinformatics: Top 10 Tools
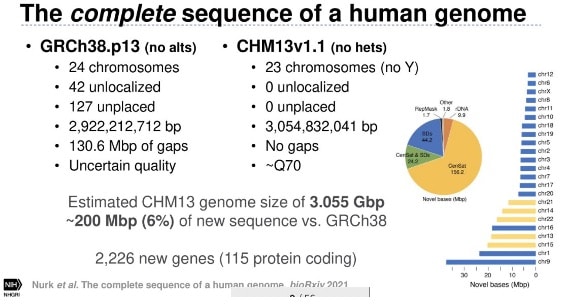
Another exciting part of the event concentrated on bioinformatics. Serendipitously, these bioinformatics sessions coincided with the Telomere-to-Telomere Consortium’s release of the first ‘complete’ human genome. In her keynote address at SMRT Leiden 2021, consortium member Arang Rhie (@ArangRhie) gave a behind-the-scenes look at how the team sequenced a human genome in its entirety for the first time ever in history. This included characterizing the final unresolved 8% of the genome.
The other keynote by Tobias Marschall (@tobiasmarschal) of Heinrich Heine University Düsseldorf provided an interesting review of haplotype-resolved assemblies and the Human Genome Structural Variation Consortium.
Still more, speakers presented the tools they’ve developed to optimize HiFi reads, including:
● Merfin – k-mer-based assembly and variant calling evaluation for improved consensus accuracy (Arang Rhie)
● PanGenie – algorithm that leverages a pangenome reference built from haplotype-resolved genome assemblies in conjunction with k-mer count information from raw, short-read sequencing data to genotype a wide spectrum of genetic variation (Tobias Marschall)
● SQANTI3 – an automated pipeline for the classification of long-read transcripts that can assess the quality of data and the preprocessing pipeline (Rocío Amorín de Hegedüs @rocioadh)
● tama (Transcriptome Annotation by Modular Algorithms) – software designed for processing Iso-Seq data and other long-read transcriptome data (Richard Kuo @GenomeRIK)
● pbaa (PacBio Amplicon Analysis) – separates complex mixtures of amplicon targets from genomic samples to cluster and generate high-quality consensus sequences from HiFi reads (Zev Kronenberg @zevkronenberg)
● bellerophon – analyzes MHC typing and other low-complexity gene amplicon data; performs allele calling while detecting polymorphic sites within the sequences and removing potential chimeric sequence variants (Yuanyuan Cheng @Yuanyuan929)
● svpack – tools for filtering, comparing, and annotating structural variant (SV) calls in VCF format (Aaron Wenger)
● JumboDB – tool for de Bruijn graph construction (Anton Bankevich @AntonBankevich)
● uLTRA – tool for splice alignment of long transcriptomic reads to a genome, guided by a database of exon annotations. (Kristoffer Sahlin @krsahlin)
● LeafGo – workflow to rapidly produce high-quality de novo plant genomes (Luca Ermini @ermini_luca)
By and large, SMRT Leiden 2021 was packed full of valuable discussion, behind-the-scenes info, and exciting revelations about the research being done via HiFi sequencing. Each session is available to watch on demand. Register for free now, before it’s too late.