Been itching to talk about your latest single-cell experiments, your favorite differentially expressed isoforms, or your latest and greatest software for visualizing alternative splicing, but thwarted by a worldwide pandemic preventing in-person scientific events? 
We were too, so we organized a virtual social club to easily enable scientists to geek out together. And we weren’t disappointed by our first event, which attracted dozens of self-proclaimed Iso-Seq analysis geeks and other curious researchers to share their work (published, unpublished and in progress) and discuss the benefits and challenges of incorporating long-read transcript sequencing into their research.
Welcome to the Iso-Seq Analysis Universe
PacBio’s own Iso-Seq analysis expert, Elizabeth Tseng (@magdoll) kicked off the Iso-Seq Social Club with an introduction to the method, which uses PacBio’s HiFi reads to characterize full-length transcript isoforms. The Iso-Seq method has been used to identify aberrant splicing in genetic diseases, characterize alternative promoter usage in cancer, and is making its way into the single-cell space for studying subregions in postnatal mouse brains and even ant brains!
But none of these studies are possible without proper tools, and as attendees learned, bioinformatics tools made specifically for long-read transcriptome data is a bustling field.
Francisco Pardo-Palacios (@FJPardoPalacios) and Ángeles Arzalluz Luque (@aarzalluz_), both from the Ana Conesa lab at Universitat Politècnica de València, presented the trilogy of SQANTI, IsoAnnot, and tappAS, which takes the output from the PacBio Iso-Seq analysis through classification, functional annotation, and differential analysis. Many of these tools are now becoming the standard workflow for Iso-Seq studies.
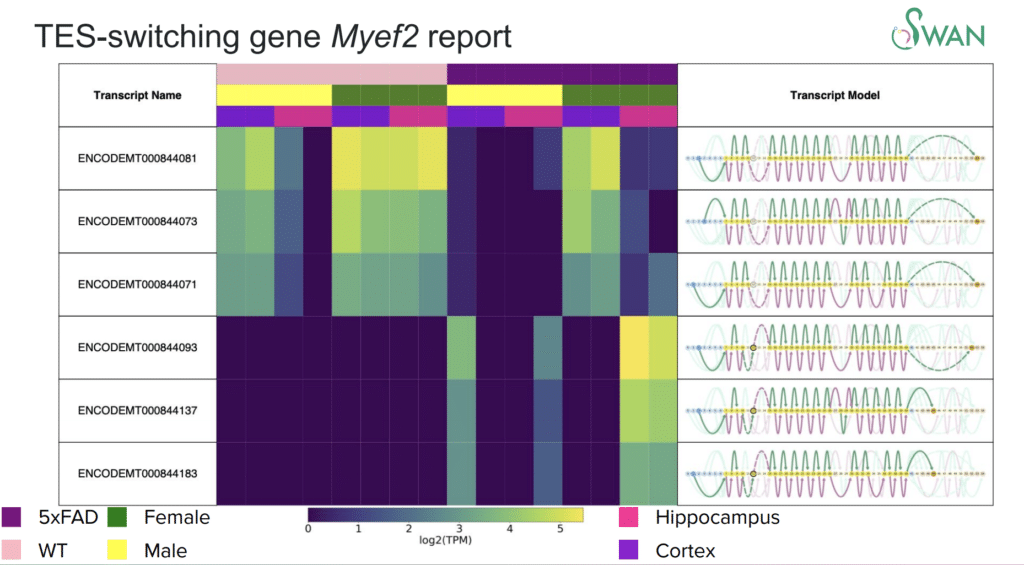
Fairlie Reese (@FairlieReese), a PhD candidate from UC Irvine, presented her tool, Swan. It provides a graphical representation of alternative splicing events, but can also be used to detect differential isoform usage and isoform switching events.
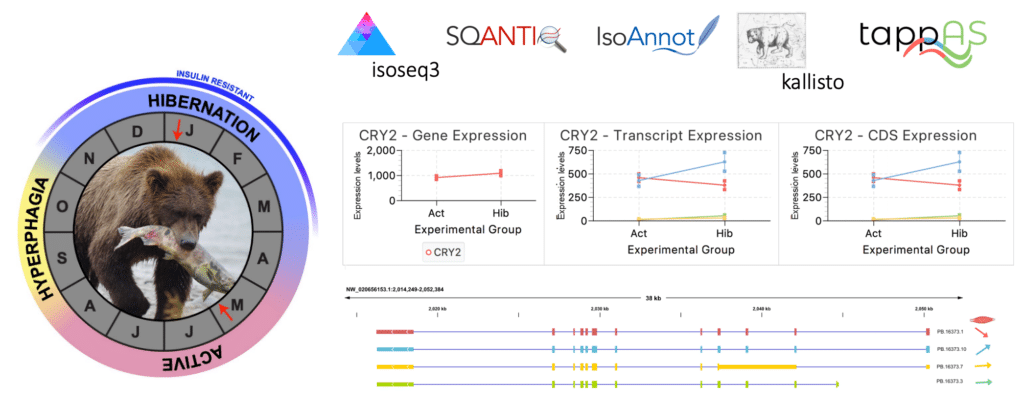
The Hunt For Differentially Expressed Isoforms In Bears… and Brains
Using Iso-Seq data on brown bears during hibernation and active seasons, Joanna Kelley (@joannalkelley) associate professor at Washington State University, was able to discover that fat tissue had higher levels of differential isoform usage (DIU) compared to liver and muscle tissues.
“Genes that show no change in expression levels but show major isoform switching and differential isoform usage are the ones we’re most interested in, because those are isoforms that we can’t quantify in any other way,” Kelley said.
Jack Humphrey (@JackHumphrey_), a postdoc in the Towfique Raj lab at Mount Sinai, is using Iso-Seq analysis to study complex splicing in genes associated with Alzheimer’s disease risk. Humphrey shared data from 30 post-mortem isolated microglia they collected. He also presented the processing pipelines for annotating and classifying the Iso-Seq transcripts, with an emphasis on filtering potential library artifacts – an often neglected but critical aspect of any bioinformatics work. Using a combination of existing tools and custom filtering, Humphrey showed that the curated transcriptome is high-quality and has already revealed interesting splicing events not observed with short-read data.
Single-Cell Iso-Seq Method for Precision Oncology and Hematopoietic Lineages
Arthur Dondi (@ArthurDondi), a PhD candidate from ETH Zurich, is using single-cell Iso-Seq (scIso-Seq) to study ovarian cancer. Specifically, by characterizing full-length isoforms in the omentum (fatty tissue covering the abdomen), there’s a potential for discovering neoepitopes and therapeutic targets.
Dondi and collaborators employed the HIT-scIso-Seq technique, which employs TSO artifact removal and concatenation for cDNA molecules coming out of the 10X single-cell platform, and increased the number of reads per SMRT Cell 8M by six-fold. They are planning to query this rich dataset for differential isoform expression, novel isoforms and fusion discovery.
Vladimir Souza from University of Zurich is working on calling variants from Iso-Seq data, showing that using DeepVariant or GATK with specific parameters achieved the highest precision-recall. The goal of his project is to eventually link the variations to changes in ORF predictions.
Anita Scoones (@AnitaScoonesPGR), a PhD candidate from the Earlham Institute, is studying lineage bias during hematopoietic stem cell differentiation. She wants to use single-cell Iso-Seq analysis on their plate-based single-cell libraries, similar to how her lab mate Laura Mincarelli had used long reads to look at isoform differences in aging mice.
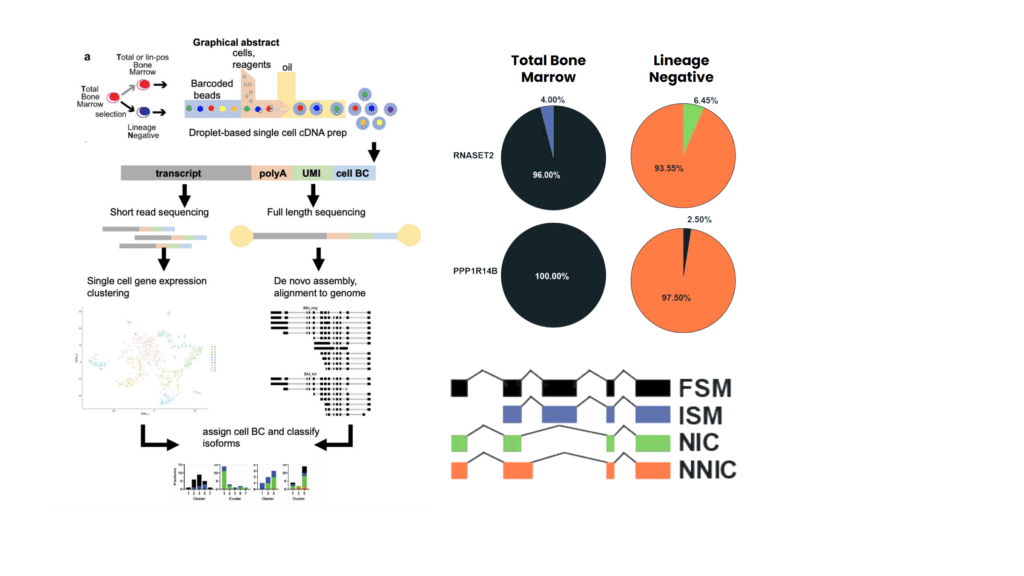
Anne Deslattes Mays (@adeslat) and Marcel Schmidt of Georgetown University had previously used bulk Iso-Seq analysis to show that lineage-negative cells in bone marrow have higher isoform complexity than lineage-positive cells. They are now pushing the question into the single-cell space: is isoform diversity uniform at the single-cell in lineage-negative cells? Applying the scIso-Seq method, they found striking differences between the total and lineage-negative bone marrow subpopulations, where lineage-negative cells had an overwhelmingly high number of novel isoforms and were enriched in spliceosome-associated genes. This suggests that alternative splicing in lineage-negative cells is attributed to cell-fate decisions of each cell subpopulation.
What’s Next For Iso-Seq Analysis?
The event ended with a lively discussion in which attendees discussed the need for bioinformatics tools that can handle large amounts of Iso-Seq data and create reproducible workflows that others can easily adapt. They also addressed the one-size-fits-all approach of using a single reference annotation and said a re-think may be in order.
“Maybe references should be qualified by the tissues or cell types of interest,” suggested Ana Conesa (@anaconesa). “How do we use all these novel isoforms to annotate the transcriptome?”
Mays agreed that “the best reference is self.”
In neuroscience, scientists have a poor idea of what makes a cell type-specific isoform, Humphrey said. The challenge is agreeing on what a definitive reference for each cell type would be, he added.
“We’re not done at just references,” Schmidt suggested. “We need to assign a function to these isoforms, even if it’s a regulatory one.” And Conesa said a system level of analysis is necessary.
Overall, the enthusiasm around Iso-Seq analysis is consistent. The promise of a properly defined transcriptome summarized the conversation and paves the way for future discussion.
Want to learn more? Register to watch an on-demand recording of the event, or check out these resources:
PacBio Applications and Workflows
RNA Sequencing with Iso-Seq Analysis
Procedure & Checklist