
Written by: Menzies Chen, Director, Product Management, Informatics
With the launch of the Revio system, PacBio is bringing a host of technologies together to increase scale for HiFi long-read sequencing, by increasing throughput and lowering costs per Gigabase of output.
HiFi sequencing was a phenomenal breakthrough when it was introduced in 2019, bringing the ability to produce highly accurate long-read information that short reads simply don’t capture. It makes us incredibly proud to have grown the usage of HiFi sequencing into so many parts of the scientific community that Nature Methods named long-read sequencing the Method of the Year for 2022.
While our Sequel IIe platform brought HiFi into the spotlight for scientists, we knew there was so much untapped potential from larger, more ambitious projects for which the economics and throughput of the Sequel IIe system was not adequate.
Challenging ourselves to deliver higher throughput and lower operating costs
We challenged ourselves to address the major barriers for broader adoption of HiFi sequencing: increase throughput and reduce the cost to enable larger-scale projects.
Our research and development teams came back with technology that could triple the number of ZMWs on a SMRT Cell, and robotics to handle 4 SMRT Cells simultaneously — yielding an immediate 12x increase in throughput relative to a Sequel IIe system. While that was exciting, it created an additional computational challenge. How much more compute would be needed to keep up with the increase in throughput, and would it explode the costs to build the instrument? Or would customers have to wait for compute to process all that data before more sequencing could be done?
The main compute task for HiFi sequencing is the process of computing a consensus read using the Circular Consensus Sequencing algorithm (CCS). Sequencing on PacBio long-read instruments produces raw reads that represent DNA polymerase processing the sample DNA which has been circularized so that polymerase can sequence multiple passes of the sample throughout the sequencing process. Compute then calculates a consensus sequence from the multiple passes, and is performed across millions of sequencing reactions taking place on each SMRT Cell. Sequencing for a longer period of time generates more passes through each molecule, and more passes produce more accurate HiFi reads.
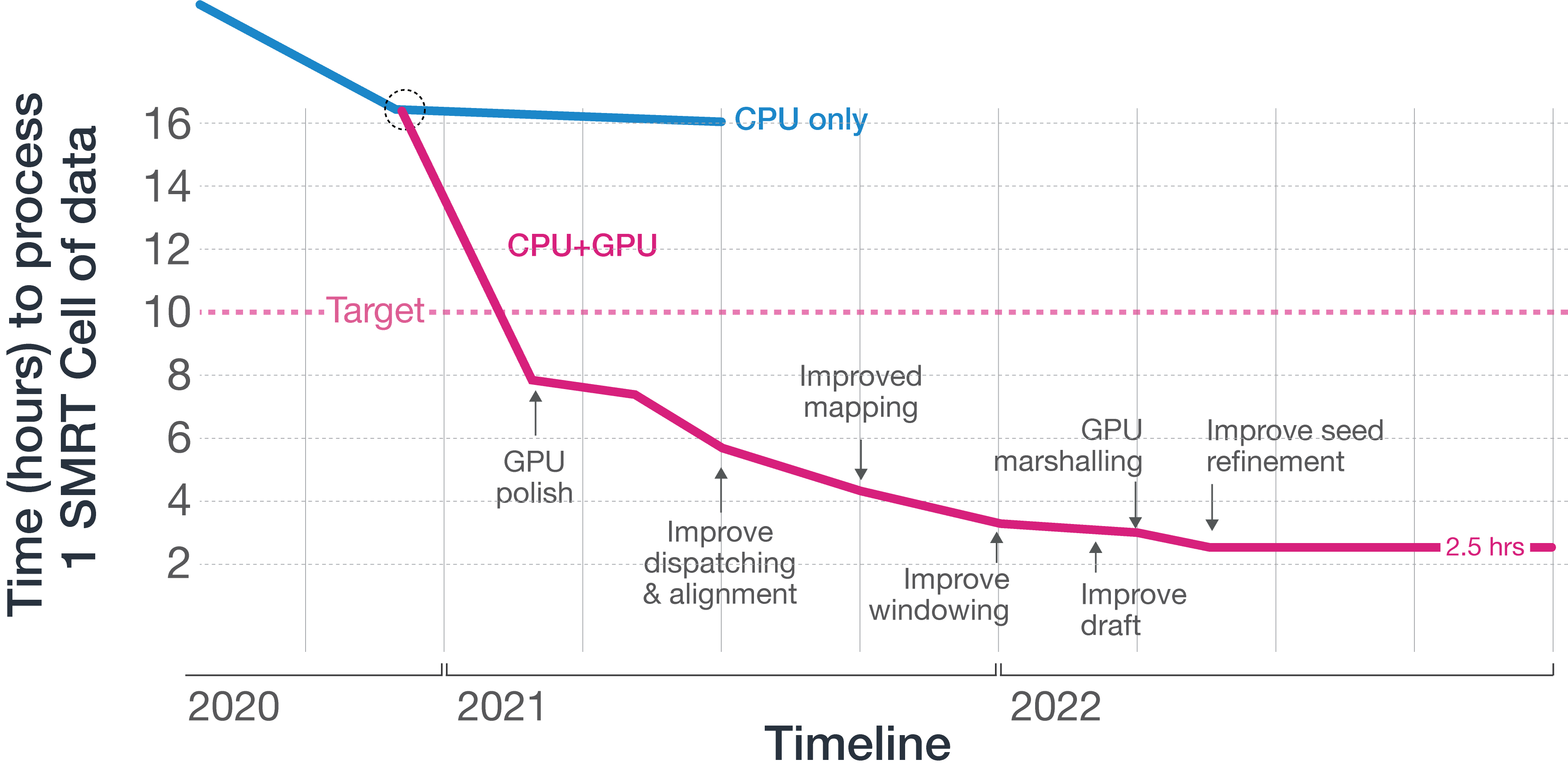
With our compute infrastructure from the Sequel IIe system, we simply were not able to meet our targets for a commercially viable product, with the CCS step taking over 16 hours per SMRT Cell on the new platform. We could fit up to two compute units into a machine without expanding upon the instrument footprint, so performing the CCS step for each SMRT Cell in under 10 hours allows us to process data from 4 SMRT Cells in the time it takes to sequence another set of SMRT Cells.
Our CPU-only architecture from the Sequel IIe platform, however, could only take us so far, so we began to explore accelerator technologies such as GPU and FPGA. Our engineering team quickly decided to develop on GPU due to the speed with which we could develop and deploy a solution for our new platform. By using advanced GPUs from NVIDIA, our team began moving workloads to GPU, starting with the GPU polish step, which had consumed over 80% of the CPU time on the Sequel IIe system. Polishing uses a hidden Markov model, for which NVIDIA GPUs are well suited, by being able to keep hot data in shared memory when filling out dynamic programming matrices. By itself, moving this step to GPU enabled a commercially viable product, but our team was able to move additional compute steps to GPU, and was ultimately able to deliver an implementation of CCS that could complete its task in 2.5 hours, generating considerable headroom for us to do even more with the newly available onboard compute afforded by adding GPU.
With the extra headroom, we then looked at ways to increase yield and accuracy through software and deep learning.
Adding DeepConsensus
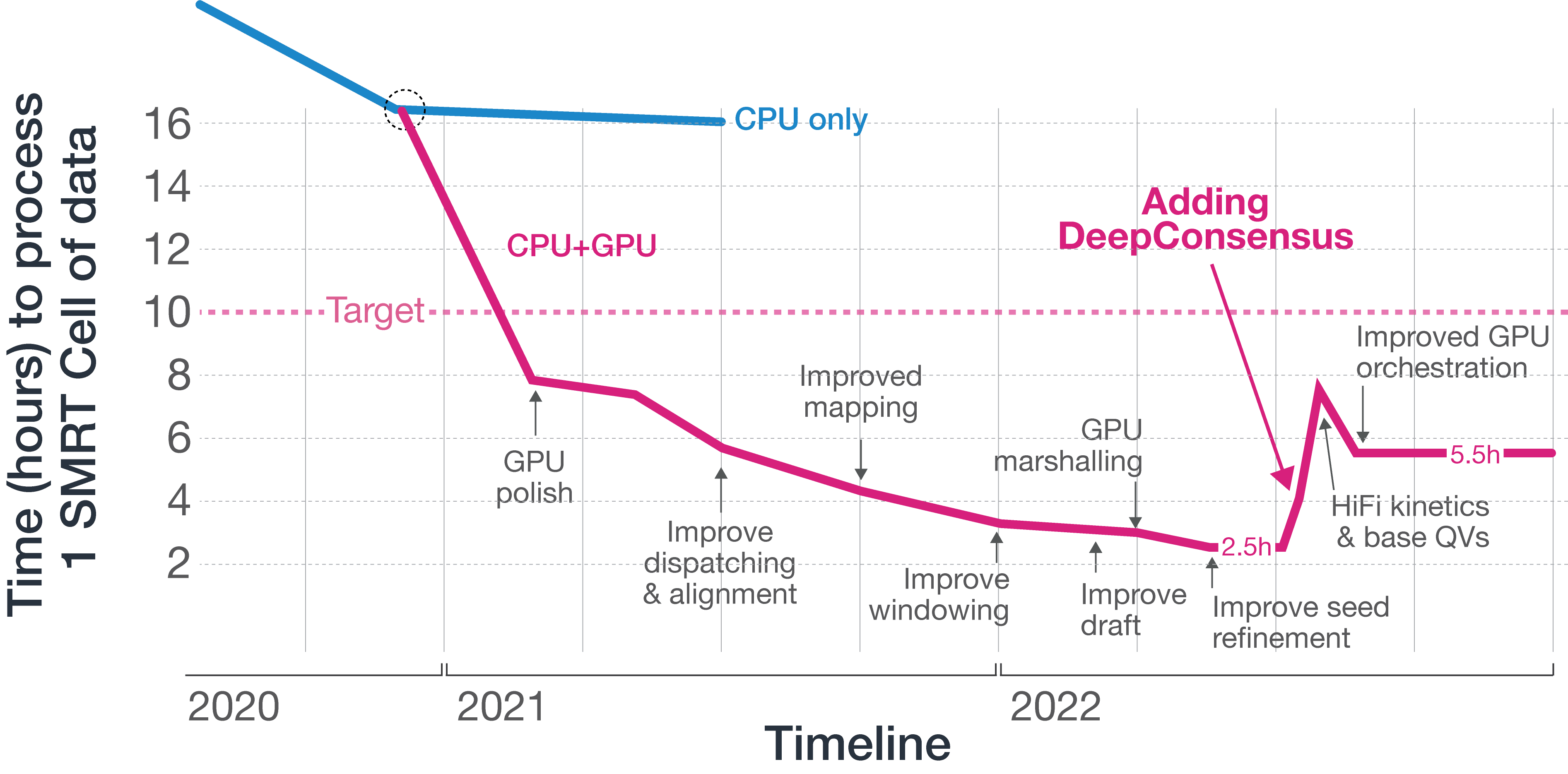
DeepConsensus is a deep-learning algorithm published by Google Health in 2021 that takes subreads from PacBio instruments and partitions them into tensor objects that are fed into an encoder-only transformer to produce a model that outputs polished segments that are stitched into polished reads. These polished reads are more accurate than subreads and HiFi reads without DeepConsensus. The original work is compute-intensive and requires storing raw read data; at the time of publication, it required 16,000 CPU hours to process a Sequel IIe SMRT Cell 8M and 500GB of data storage space to store raw read information. Assuming a 12x increase in data throughput from a putative new platform, this would have resulted in 192,000 CPU hours and 6TB of storage per SMRT Cell.
We are fortunate to have established a fantastic collaboration with Google Health to investigate building DeepConsensus into our new platform, and using our compute headroom to deliver this capability to our customers.
Since publication, Google has made significant model improvements to speed up the time it takes to process data from the Sequel II and IIe systems, and our engineers have been able to build on that work and optimize its implementation on NVIDIA GPUs. We have also given Google access to early data from the new platform to re-train their model, and the result is an amazing outcome for PacBio customers.
Adding DeepConsensus extended the time to process each SMRT Cell on our new platform, ultimately landing around 5.5 hours per SMRT Cell, per GPU. Being able to achieve higher accuracy using DeepConsensus now allows us to deliver accurate HiFi reads to customers in a shorter amount of time; whereas the Sequel IIe has a standard sequencing time of 30 hours, for our new platform, we can now reduce that to 24 hours.
Delivering Revio
The result is the Revio system, and we’re so excited to get this into our customers’ hands! In addition to our development teams and our collaborations with Google Health and NVIDIA, we’re proud of the work we’ve done to implement DeepConsensus on NVIDIA GPUs in Revio.