
Written by: Menzies Chen, Associate Director, Product Management, Informatics
Whether it is accessing areas of the human genome that short reads can’t sequence, or powering genome assemblies across all of biology, at PacBio, we’re continually looking for ways to enable our customers to discover more using our long-read sequencing technology.
Today, we’re excited to share the latest milestone in this pursuit with the launch of SMRT Link v11.0. The new release is available for download now, unlocking an exciting menu of new capabilities and updates, including:
Support for SMRTbell prep kit 3.0
SMRT Link has been updated to support the new SMRTbell prep kit 3.0. This reduces workflow time by up to 50% and input DNA requirements by up to 40%, down to 3 micrograms per human genome or as little as 300 nanograms when multiplexing many smaller genomes. SMRT Link now also reflects the new branding PacBio introduced in February.
5mC calling: accessing a new dimension in genomic data
SMRT Link v11.0 adds a new capability to directly measure 5-methylcytosine (i.e. DNA methylation) at CpG sites from native DNA samples. By activating 5mC calling within SMRT Link Run Design, Sequel II and IIe systems will now output HiFi data with tags in the .bam file that specify the positions and probabilities of 5mC methylation at CpG sites. The added BAM tags increase the size of the HiFi data by only ~5%, and require no additional modifications to current workflows other than utilizing the option in the updated software where it is on by default for WGS applications. 5mC calling can also be performed on data generated in prior versions of SMRT Link with kinetic data.
Heteroduplex filtering
Heteroduplexes are DNA molecules where the forward and reverse strands are not perfect reverse complements. With SMRT Link v11.0, users can now automatically detect heteroduplexes in a SMRTbell library to output sequences from each strand separately. Heteroduplex detection is an improved replacement for by-strand mode used with previous versions of SMRT Link. This option is available for Adeno-Associated Virus, Full-Length 16S rRNA Sequencing, or either of our Amplicon applications in Run Design.
Delivering HiFi data by default
As we build upon the high-accuracy data that HiFi sequencing provides, we are refining how data are packaged when coming off our instruments.
In SMRT Link v11.0, Sequel IIe instruments will now output HiFi data by default by generating a *.hifi_reads.bam file for each SMRT Cell that contains only HiFi reads (Q20+). Users may access low-quality reads using a parameter in Run Design. Users who rely on subreads for their workflows will still be able to access them by importing a run design CSV, as described in our updated user guide. On Sequel II instruments, circular consensus sequencing to produce HiFi reads will be performed automatically through SMRT Link. This simplifies the need to specify where CCS processing occurs and removes the need to store large volumes of subreads data.
Instrument view in SMRT Link
SMRT Link v11.0 now provides a view of connected instruments so that instrument status and real-time metrics are available within SMRT Link.
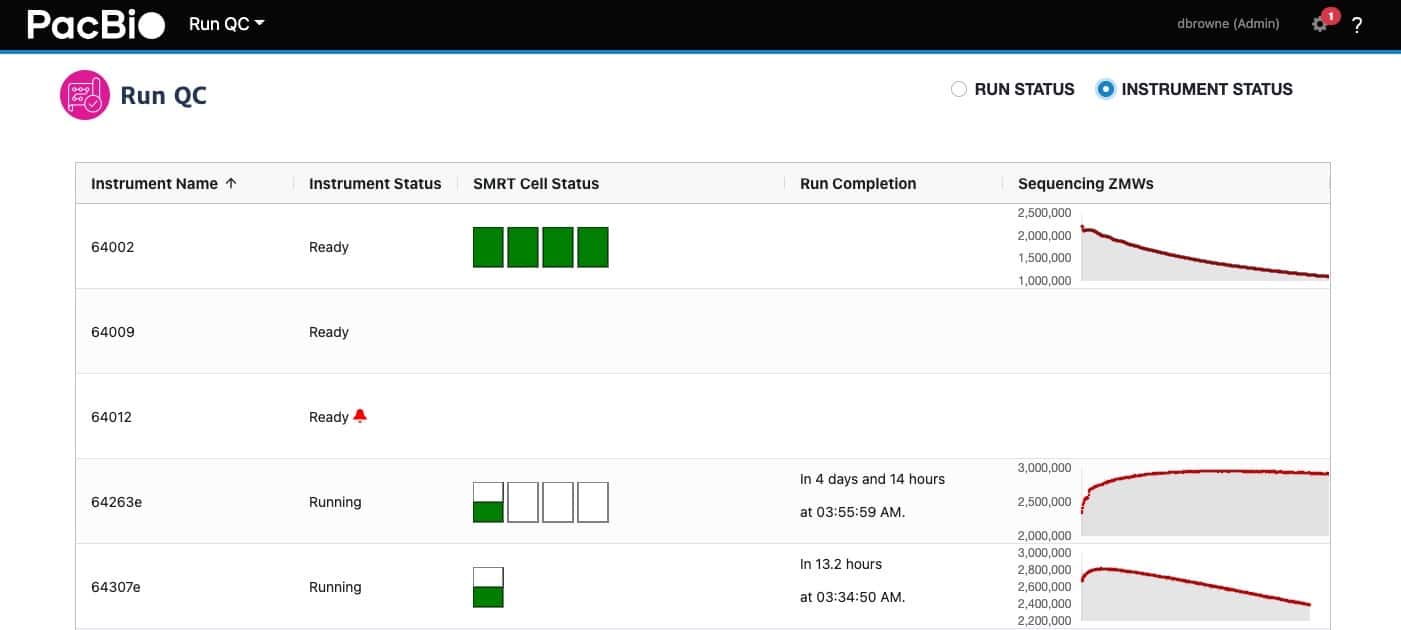
The SMRT Cell status indicator shows whether SMRT Cells on Sequel II and IIe instruments are in progress or completed, as well as error states. The Run Completion column shows the current estimate for when the run will complete, and a new Sequencing ZMWs view shows a graph of active zero-mode waveguides on the SMRT Cell that are currently sequencing.
SMRT Link Sample Setup HT
When calculating the volumes needed to prepare a sample through the Annealing, Binding, and Cleanup step, SMRT Link v11.0 also now has a High-Throughput mode that will output instructions to align with how users process multiple samples together. This mode produces instructions for producing a larger volume of master mix that is then portioned and combined with each sample.
That’s a wrap on the highlights for this release, with a lot more happening under the hood. Stay tuned for future posts that will highlight new features and capabilities in our software, and shed some light into design decisions from the product team. If you have any comments, questions, or suggestions, please reach out to us at SWProduct@pacificbiosciences.com, and subscribe for more in-depth posts about our new features, products, and releases.