
We hear a lot about the growing crisis of antibiotic resistance in human health, but it turns out this is just the most visible place it appears as it moves through our complex modern environment. For example, when intensive farming is used to feed large urban populations, antibiotic resistance can first emerge on farms and gain access to human communities through the food system.
One of the key groups on the front lines of monitoring antibiotic resistance from farm to fork in the United States is the National Antimicrobial Resistance Monitoring System (NARMS). NARMS was launched in 1996 as an interagency partnership among the USDA, the FDA, the CDC, and state and local health departments to protect public health by tracking changes in the antimicrobial susceptibility of bacteria in food animals, retail meat and ill people.
Nationwide, public health labs submit Salmonella, Campylobacter, Shigella, Escherichia coli O157, and Vibrio isolates from clinical specimens and outbreaks to the CDC for testing. In addition, 19 states collect samples of retail chicken, ground beef and pork chops every month for culturing, serotyping, antimicrobial susceptibility testing, and genome sequencing by the FDA. Finally, the USDA conducts similar tests on bacteria isolated from food animals at randomly sampled, nationally representative slaughter and processing plants throughout the country.
By combining information from all these sources, NARMS can detect emerging trends in resistance, understand the genetic mechanisms of resistance, link illnesses to specific sources or practices, educate consumers, and develop data-driven recommendations for improving antibiotic stewardship.
One of the tools NARMS uses for bacterial whole genome sequencing is PacBio long-read sequencing, prized for its ability to assemble not only chromosomes but also plasmids and other accessory genome elements that frequently carry drug resistance genes. Over the years, scientists at NARMS have used PacBio reference genomes to facilitate numerous comparative genomics analyses of Salmonella, Campylobacter and Enterococcus strains, examining how virulence and resistance to β-lactamase, ciprofloxacin, linezolid and other families of antibiotics evolves at the molecular level.
Adding to this body of work, NARMS Director Patrick McDermott and collaborators recently reported applying SMRT Sequencing to 11 E. coli isolates collected from retail meats. One of the explicit goals of the study was to add more closed plasmids carrying quinolone resistance to their reference database, expanding our understanding of this emerging challenge to the treatment of Gram-negative infections. All the selected E. coli strains used in this study were resistant to ciprofloxacin and known to carry plasmid mediated quinolone resistance (PMQR) elements. The team generated “closed, circular chromosomes and plasmids from each isolate,” they write.
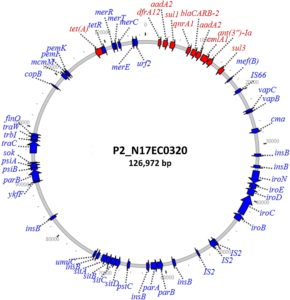
One key finding of the study was that seven of the plasmids analyzed did not match any existing sequences in GenBank. The authors commented, “This demonstrates the importance of increased sequencing of plasmids even in well-studied bacteria such as E. coli, since completely new plasmids are still being discovered.” They also note that while the prevalence of PMQR genes in the US food supply is currently low and the E coli strains from this study are unlikely to cause disease in humans, the identification of numerous novel plasmids suggests the potential for further spread to other strains or genera.
Furthermore, the authors emphasized that “This work shows the value of long-read sequencing in de novo characterization of [antimicrobial-resistant] plasmids.” More specifically, “Using only short-read sequencing data makes it difficult to accurately identify plasmids or fully characterize them.”
For example, closed plasmids are required to identify when multiple resistance genes or multiple copies of the same gene are co-located in one plasmid. This more complete information is important for uncovering the potential for co-selection of resistance. Another key finding from the paper is that while fluoroquinolone is not commonly used in food animal production, seven out of the 11 PMQR plasmids sequenced in this study also carried genes for resistance to tetracycline, “the highest selling antimicrobial for food animals in the United States.” Continued use of tetracycline in food animals could therefore drive co-selection for fluoroquinolone resistance in E. coli.
Learn more about the methods and workflow for bacterial whole genome sequencing.