With Earth Day almost here, it is important to remember that our planet is made up of countless interconnected parts, from vast oceans to the microscopic world of microbes. Despite their tiny size, microorganisms play a massive role in shaping our world. They have a hand in everything from cloud formation to food production, medicine, and potentially even human behavior. Given this seemingly outsized influence on ecological processes, scientists at Michigan Technological University are now looking to bacteria to aid in the fight to solve some of the biggest environmental and humanitarian challenges of our time: plastic waste and food insecurity.
Plastic breakdown directly by microorganisms is often a slow and inefficient process because plastic polymers take a great deal of energy to break apart. However, chemical methods for plastic deconstruction can rapidly break plastics down into intermediate compounds that microorganisms can process and convert into biomass more completely.
An innovative approach to solve a challenging problem
Knowing these difficulties, Stephen Techtmann’s research group at Michigan Tech is developing a chemical and biological process for the conversion of plastic into food. This flexible system involves the conversion of olefin plastics (HDPE, LDPE, PP) and ester-based plastics (PET) into biomass using two chemical processes and a consortium of bacteria that can grow on plastic-based inputs. The bacterial biomass cultivated in this process could be recovered as microbial cells and used as a food or food supplements (e.g., single cell protein) in ways similar to nutritional yeast or Spirulina. Early in their study, the Techtmann lab realized that to optimize and increase the growth efficiency of the biomass within their system, they needed a way to confidently identify and extensively characterize unique species and strains within the plastic degrading microbial communities they developed.
Kickstarting progress with PacBio long-read metagenomics
Before being awarded a PacBio SMRT Grant in 2021, the researchers only had conventional short-read data on the microbial communities used to digest and flexibly convert plastics. This left the team with an incomplete picture of the composition and collective functional capabilities of their bacterial consortia. So, to fill this information gap and advance their process design they utilized PacBio long-read metagenomics to successfully capture, and close metagenome assembled genomes (MAGs) for strains found within their experimental microbial communities.
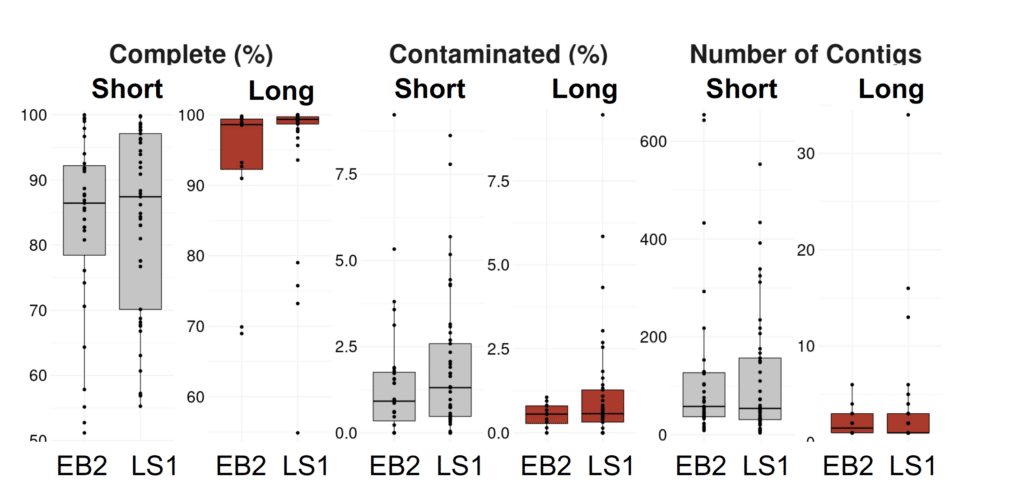
The team’s ability to identify genes and annotate functions more effectively with HiFi MAGs has helped them understand which organisms perform specific metabolic functions in plastic deconstruction and enabled them to tease out details like division of labor and specialization within the community.
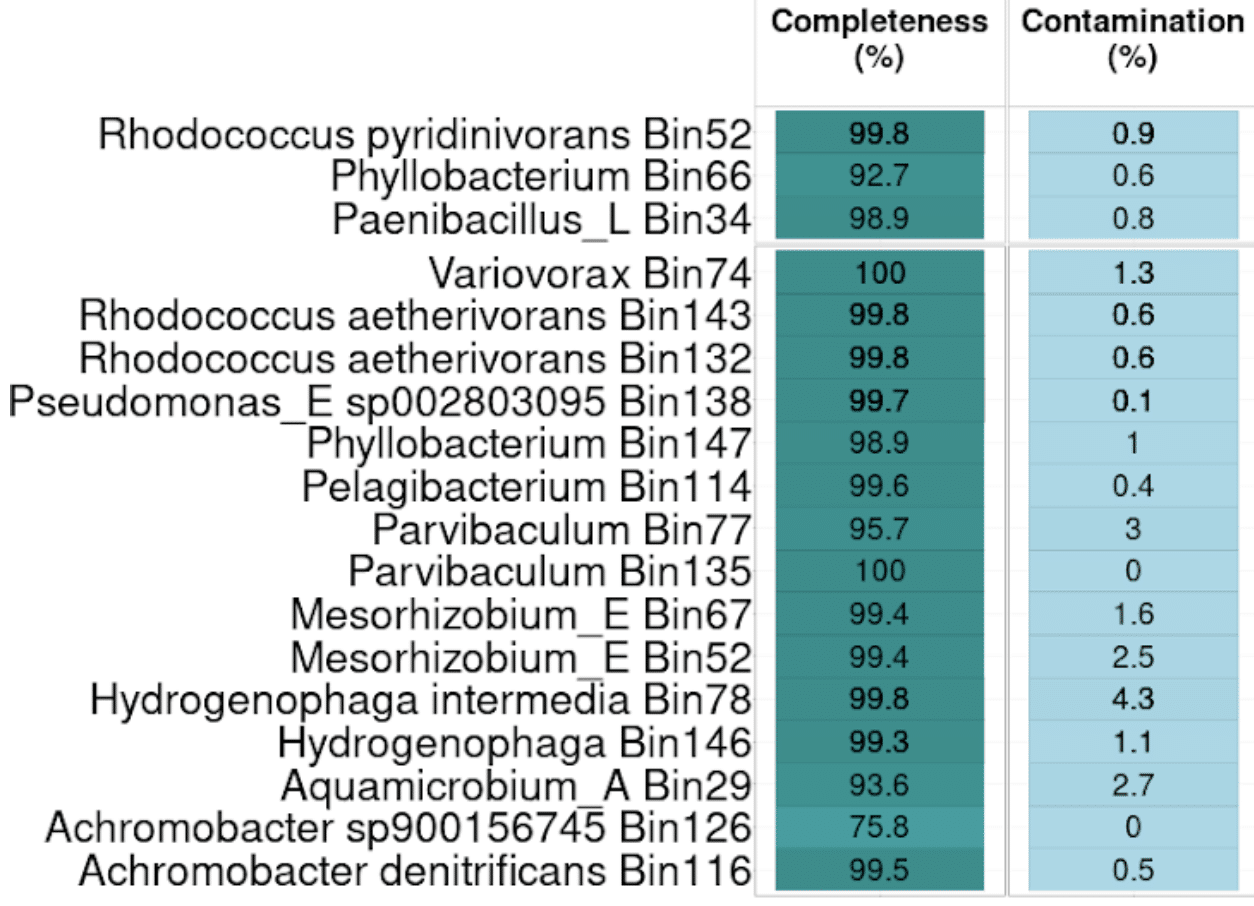
Now that access to a clearer picture of the microbial community structure has been unlocked using PacBio HiFi metagenomics, Techtmann’s lab is focusing on ways to target and optimize more granular aspects of biomass production from plastic waste. Building on this success, the team has begun to plan out a pipeline for examining the safety and nutritional content of the microbial biomass through chemical analyses, whole animal screening, and in silico screening for genes associated with the production of toxins and allergens. They hope that this, coupled with chemical and biological processes will make it possible to generate a low-cost dietary supplement that could revolutionize the way people all over the world combat hunger, while also working towards a cleaner environment by reducing plastic waste.
Lead the way to a brighter future in balance with our planet
Sequencing technology is entering a new era, one where the read length and accuracy necessary to realize the full potential of microbial research can be accessed faster and more affordably by more research teams around the world than ever before. Despite this, the microbial presence in our daily lives remains both profound and still largely unknown. To make a real impact, it will take enterprising minds with unprecedented access to information about microbial genomes and community profiles to make a difference. In metagenomics, HiFi sequencing enables researchers to not only forge ahead and explore entirely new avenues of discovery but also reexamine existing challenges with important implications for how humanity can more sustainably interface with our living world.
Let us all work together to deliver the promise of genomics to build a brighter future for the earth and for ourselves.