UPDATE: This paper has now been published in HGG Advances from Cell Press
In an exciting new preprint, scientists from the HudsonAlpha Institute for Biotechnology and the University of Alabama at Birmingham describe the use of PacBio highly accurate long-read sequencing to identify pathogenic variants responsible for previously undiagnosable, rare neurodevelopmental disorders.
Lead author Susan Hiatt (@suzieqhiatt), senior author Gregory Cooper, and collaborators conducted genomic analyses of several family trios in an attempt to find causal genetic variants that had been missed with earlier studies.
“Large fractions of [neurodevelopmental disorders] cannot be attributed to currently detectable genetic variation,” they report. “This is likely, at least in part, a result of the fact that many genetic variants are difficult or impossible to detect through typical short-read sequencing approaches.”
The project involved six family trios with children affected by neurodevelopmental disorders who had previously had their genomes sequenced with short-read technology. In all cases, “no causal genetic variant, or even potentially causal variant, was found,” the scientists write.
To test their idea that the disease-causing variants were missed by short-read sequencing platform, they next turned to PacBio for long-read whole genome sequencing. Using SMRT Sequencing, on a Sequel II System, they generated HiFi reads (highly accurate long reads) that were each >99% accurate.
HiFi results “were used to detect variation within each trio and generate de novo genome assemblies, with a variety of metrics indicating that the results are more comprehensive and accurate, especially for complex variation, than those seen in short-read datasets,” the authors note. “Detection of simple-repeat expansions and variants within low-mappability regions, for example, was far more accurate in [HiFi] data than that seen in [short reads], and many complex SVs were plainly visible in [HiFi] data but missed by [short reads].”
For all six trios, SMRT Sequencing was performed in HiFi mode on the Sequel II System, covering each proband genome to an average of 30x and each parent’s genome to an average of 16x. An analysis of structural variants — the type of variation most likely to be missed by short reads — found that each trio collectively had about 56,000 structural variants, with an average of nearly 60 candidate de novo variants per child.
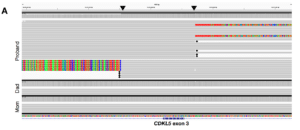
For two of the probands, the results got even better. In one case, the team identified a de novo heterozygous insertion of nearly 7,000 bases in an intron of the CDKL5 gene that they deemed likely pathogenic. Since CDKL5 has been associated with early infantile epileptic encephalopathy 2, a condition characterized by many symptoms connected to this proband’s case, “we prioritized this event as the most interesting candidate variant,” Hiatt et al. report. “To determine the effect of this insertion on CDKL5 transcripts, we performed RT-PCR from RNA isolated from each member of the trio.” Results supported their theory that the variant has a loss-of-function effect for the individual.
For the other proband, the team found a de novo structural variant that affected two genes, DGKB and MLLT3. “[HiFi] reads and contigs from the proband’s de novo assembly support the existence of at least three breakpoints, suggesting that a ~250 kb fragment harboring three coding exons of DGKB are removed from chromosome 7 and inserted into an intron of MLLT3 on chromosome 9,” the scientists report. A qPCR analysis demonstrated that the proband had less than two-thirds the expression of MLLT3 than her parents or unrelated individuals. The variant, while intriguing, was classified as a variant of uncertain significance.
“The breadth and quality of variant detection coupled to finding variants of clinical and research interest in two of six probands with unexplained [neurodevelopmental disorders] strongly support the value of long-read genome sequencing for understanding rare disease,” the scientists conclude. “Further, as [HiFi reads] can capture complex variation in addition to essentially all variation detectable by short-read sequencing, it is likely that it will become a powerful front-line tool for research and clinical testing within rare disease genetics.”
See additional examples of the use of SMRT Sequencing in rare disease research and learn more about structural variant detection:
Webinar: Increasing Solve Rates for Rare and Mendelian Diseases with Long-Read Sequencing
The Pathologist: Solving Rare Disease with SMRT Sequencing
A Rare Opportunity to Help Tackle Daughter’s Rare Disease
Review: Long-Read Sequencing Helps Uncover Genetic Basis for Rare Disease
SOLVE-RD Team Adopts PacBio Sequel II System to Solve Rare Diseases
August 17, 2020 | Neurogenomics