The study of genomics has revolutionized our understanding of science, but the field of transcriptomics grew with the need to explore the functional impacts of genetic variation. While different tissues in an organism may share the same genomic DNA, they can differ greatly in what regions are transcribed into RNA and in their patterns of RNA processing. By reviewing the history of transcriptomics, we can see the advantages of RNA sequencing — using a full-length transcript approach — become clearer.
Reaching for the transcriptome
Even before genome sequencing became commonplace, scientists were able to measure gene expression activity using hybridization approaches like northern blots, or later, microarrays. While these techniques provided a rigorous method for RNA measurement, they were limited in that they could only measure known transcripts. Additionally, many of these methods made the subtle differences between transcript isoforms difficult to decipher.
As next-generation sequencing (NGS) grew, researchers had the ability to interrogate the activity of all expressed genes including those that were previously unknown. This was a huge leap forward in the ability to characterize the transcriptome in an organism. The approach, known as RNA-seq, quickly replaced microarrays in many labs due to the enormous amount of information it could generate.
Understanding gene activity — not just the genes encoded by the genome, but the ones that are turned on at a specific time or in a specific cell or tissue — is critical for elucidating biological consequences. But even as RNA-seq expanded, scientists grew concerned about the accuracy of results when NGS short reads had to be pieced back together computationally to create whole transcripts. This is especially difficult in complex eukaryotes where one gene can generate many isoforms that differ in their transcription start site and alternative RNA processing.
In a Mendelspod interview, Stanford genomics expert Mike Snyder (@SnyderShot) described the RNA-seq process this way: “We take RNA, we blow it up into little fragments, and then we try and assemble them back together to see what the transcriptome looked like in the first place… You can’t always figure out which parts of the puzzle belong together.”
A similar concept was presented by Ian Korf (@IanKorf) in a Nature Methods article, where he equated RNA-seq to reassembling magazine articles that had been put through a shredder. Suffice it to say, there have been concerns about the need for extensive computational inference when using an RNA-seq approach.
Capturing the whole transcriptome with the advantages of long reads
What scientists really needed went beyond RNA-seq; for the most accurate view of biology possible, they had to have a way to capture isoforms in their entirety. The introduction of long-read sequencing answered this need.
With Single Molecule, Real-Time (SMRT) sequencing, researchers can generate highly accurate long reads, or HiFi reads, that are tens of kilobases long. That’s enough to capture most transcripts in a single sequencing read, with no downstream bioinformatic assembly required. This became known as the Iso-Seq method because it enabled full-length isoform sequencing, which can be used to explore a transcriptome in its entirety or in a targeted fashion to examine individual genes.
As scientists began using SMRT sequencing for transcriptome projects, a common theme emerged: long-read studies, even of the most well characterized organisms, routinely showed far more transcript diversity than previously observed. It quickly became clear that short-read NGS platforms greatly underestimated the diversity of expressed isoforms. Key published examples of the Iso-Seq method revealing previously unknown isoform diversity across different types of organisms include:
- Revealing the transcriptomic complexity of switchgrass by PacBio long-read sequencing
- Full-length transcriptome analysis of Litopenaeus vannamei reveals transcript variants involved in the innate immune system
- Single-Molecule Real-Time (SMRT) full-length RNA sequencing reveals novel and distinct mRNA isoforms in human bone marrow cell subpopulations
Beyond alternative splicing
In addition to alternative splicing, the Iso-Seq method is important for understanding alternative polyadenylation, genome annotation, fusion transcripts, isoform variant phasing, and long noncoding RNAs. Here are just a few examples of how the Iso-Seq method allowed for new transcriptome studies:
Alternative polyadenylation
Connect alternative polyadenylation motifs to specific isoforms to determine their role in agronomically important traits.
Genome annotation
Gain a complete view of the porcine transcriptome including the identification of genes missing in the assembly.
Warr, A., et al. (2020) An improved pig reference genome sequence to enable pig genetics and genomics research. GigaScience, 9, 1-14.
Fusion transcripts
Discover novel fusion transcripts in clinically relevant genes to understand disease mechanisms.
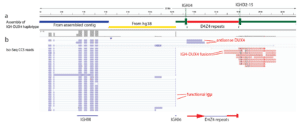
Tian, L., et al. (2019). Long-read sequencing unveils IGH-DUX4 translocation into the silenced IGH allele in B-cell acute lymphoblastic leukemia. Nature Communications, 10(1), 2789.
Isoform variant phasing
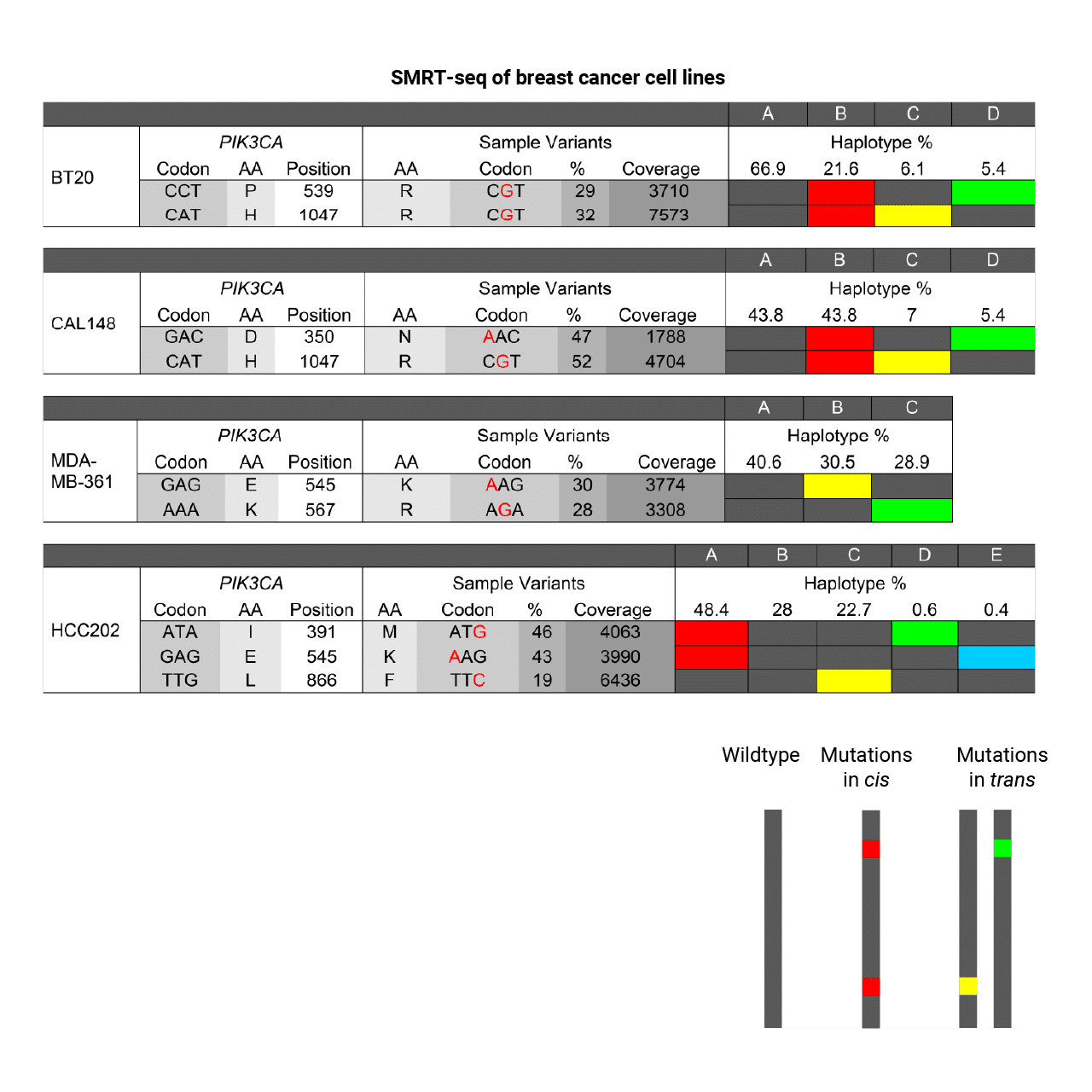
Detect and phase mutations in disease genes for clinically relevant insights.
Long noncoding RNAs
Identify long noncoding RNA for a more comprehensive view of transcriptional and translation regulation
Teng, K., et al., (2019) PacBio single-molecule long-read sequencing shed new light on the complexity of the Carex breviculmis transcriptome. BMC Genomics, 20(1), 789.
Full-length transcript sequencing offers many benefits to scientists looking to interrogate transcriptomes. Whether exploring the human transcriptome to better understand health and disease or studying plants and animals to advance agricultural or conservation efforts.
Learn more about how discovering full-length transcripts could make a difference for your research and contact a PacBio scientist to get started with your next sequencing project.
Explore other posts in the Sequencing 101 series:
The evolution of DNA sequencing tools
Introduction to PacBio sequencing and the Sequel II system
From DNA to discovery — the steps of SMRT sequencing
Why are long reads important for studying viral genomes?
Looking beyond the single reference genome to a pangenome for every species
Understanding accuracy in DNA sequencing
Ploidy, haplotypes, and phasing — how to get more from your sequencing data
DNA extraction — tips, kits, and protocols
Video: Sequencing 101: how long-read sequencing improves access to genetic information