Geneticists often point out that a human does not have “a” genome but rather two genomes: one inherited from the mother and another from the father. The number of complete sets of chromosomes in each cell — or haplotypes — is referred to as ploidy. Humans and most other animals are diploid (2N), having two sets. Many plants have higher ploidy; for example, the hexaploid (6N) California redwood has six copies of each chromosome.
The number of chromosome pairs not only increases the total amount of DNA in a genome, but it also increases the complexity of the genome — by increasing the number of alleles, or alternate forms of genes. Although the majority of the sequence between paired chromosomes are identical, it’s the differences that provide the breadth of biological variation within a species.
Phasing haplotypes to get a complete picture of genetic variation
Whether sequencing a giant polyploid or diploid, the goal remains the same: to get a complete and accurate representation of each copy of the genome or region of interest. This is often achieved by assembling a haploid (single copy) genome and then identifying variants, locations where the alleles differ. Many well-studied organisms, like humans, have standard haploid references against which other individuals are compared.
But identifying variants does not provide the complete sequence of the genome. That requires phasing, or determining which variants are from the same copy of a chromosome (in “cis”) and which are from different copies (in “trans”). One approach to phasing is to use mother-father-child trios: variants in the child’s genome that that are only present in one parent must be on the same chromosome. A second approach is population inference, which deduces that variants often seen in the same people are likely in phase. Both trio and population phasing are imperfect, as they require additional information and are only able to phase some variants.
Recent advances in DNA sequencing technology and the tools used to assemble and phase genomes allow large blocks of the sequence to be phased directly from DNA sequencing reads of one individual. Highly accurate long reads, known as HiFi reads, are uniquely suited to phasing haplotypes as they provide the high accuracy needed to detect single nucleotide variants (SNVs) and the read length to connect these variants over a long range.
Using HiFi reads, either alone or in combination with other technologies like Hi-C and Strand-seq, scientists have been able to produce phased genome assemblies of the rose — a complex tetraploid; the California redwood; and humans, including on of Puerto Rican decent, and one of Korean decent, and a cognitively healthy supercentenarian. The phased genomes have each provided novel insights into functionally important variants.
Phasing genes to identify allelic configuration of variants
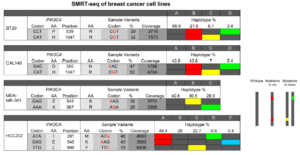
Scientists analyzing variants in the PIK3CA oncogene found a compound mutation — a double mutation that appears to give breast cancer patients an overwhelmingly positive response to the targeted PI3Kα inhibitor alpelisib. By sequencing and phasing the entire gene, the researchers were able to show that having both variants on the same allele (cis) led to a super-responder phenotype; when those variants were on separate alleles (trans), that was not the case. This information will have clinical relevance for many cancer patients and would never have been known without the ability to phase sequence data.
For recessive disease genes, it also is critical to know whether two variants seen in a gene are in trans (thus breaking both copies of a gene) or cis (thus leaving one copy intact). For example, in the case of a 9-year-old boy with multiple types of cancer, phasing of the MSH6 gene revealed that both maternal and paternal alleles carried mutations resulting in constitutive mismatch repair deficiency syndrome.
Haplotype phasing to explore the genetic origins of species
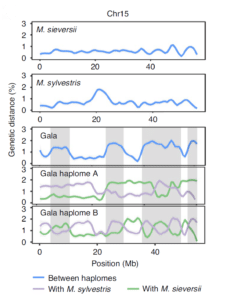
Researchers exploring apple domestication used haplotype-resolved assemblies of cultivated and wild species to better understand the genetic history of the crop. They were able to sequence and assemble full “haplomes” (haploid genomes) and showed high levels of heterozygosity with more than 20% of the Gala apple genome containing alleles derived from different wild progenitors, showing the Gala was hybrid in origin. Further, they found that introgression of new genes and alleles was a critical component to the domestication of the cultivar. This information provides better understanding of trait variability and will assist in efforts to breed for desirable traits like fruit weight and sweetness.
Allele phasing to resolve variants missed by short reads
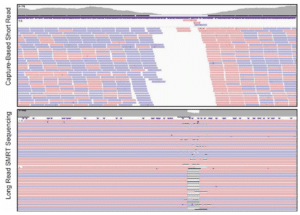
Scientists assessing the role of the promoter of the SLC6A4 gene that is thought to play a role in psychiatric disorder susceptibility, found long-read sequencing critical for interrogating a low-complexity repeat region. The length of a repeat in the gene’s promoter affects gene expression levels. Phasing the repeat length with variants in the coding region of the gene indicates whether a coding variant will have high or low expression. The authors found the repeat region was missed by short read approaches; long-read sequencing both characterized the repeat and unambiguously phased clinically significant variants that may improve pharmacogenetic testing.
How to obtain phasing information with HiFi reads
Now that you’ve seen how phasing can provide valuable insights, here is how to obtain phasing information:
- Sequence an individual with HiFi reads, which have the accuracy needed to resolve differences and the long read length to phase large haplotype blocks.
- Use a diploid-aware assembler like IPA, hifiasm, or HiCanu for genome assembly.
- Detect variants with an accurate variant caller like Google Deep Variant and then phase haplotypes with WhatHap.
- Combine HiFi data with additional technologies to extend haplotype phasing to the chromosome scale. HiFi data in combination with Hi-C or Strand-seq can phase entire genomes. If a family trio sample is available, short read data from the parents can be used to separate HiFi reads into parental bins before genome assembly (HiCanu, or during genome assembly).
To learn more about how phasing could make a difference for your research, contact a PacBio scientist to get started with your next sequencing project.
Explore other posts in the Sequencing 101 series:
The evolution of DNA sequencing tools
Introduction to PacBio sequencing and the Sequel II system
Why are long reads important for studying viral genomes?
What’s the value of sequencing full-length RNA transcripts?
Looking beyond the single reference genome to a pangenome for every species
Understanding accuracy in DNA sequencing
From DNA to discovery — the steps of SMRT sequencing
DNA extraction — tips, kits, and protocols