
Although relatively small in size, at 400-750 Mb, with seven chromosomes, the cells of roses have multiple sets of chromosomes beyond the basic set. And these can vary widely between the commercial varieties. Some are diploids, with two homologous copies of each chromosome (like humans, with one from the mother and one from the father), while others can have as many as five different sets (pentaploids). Most are tetraploids, with four sets of chromosomes.
To further complicate things, many roses are “segmental allotetraploids,” which means that part of the genome is behaving like an allotetraploid (with four chromosome sets from two distinct species, which occurs during hybridization) – and part of the genome is behaving like an autotetraploid (with four sets of homologous chromosomes).
Needless to say, parsing all of this out is challenging. But researchers from the Netherlands recently presented their solution, using HiFi reads generated by the Sequel II System.
In a workshop discussion at PAG XXVIII, Bart Nijland (@bart3601) of Genetwister Technologies (@genetwister), explained how his team set out to make a haplotype-aware assembly of Rosa x hybrida L. in order to capture its full range of genetic variation, rather than rely on more traditional assemblies which collapse the haplotypes into single sequences that could be missing critical information.
“For a highly heterozygous, highly complex, commercially important species like the rose, there is a huge benefit to making a haplotype-aware assembly,” Nijland said. “A lot of the existing technologies don’t perform very well in doing this. So we were very happy when PacBio released its HiFi protocol. Due to the high accuracy of the reads, we thought this could really help us in solving this challenge.”
The next challenge was isolating DNA from the leaf tissue of a tetraploid rose variety, which is notoriously difficult because of secondary metabolites. Once that was overcome and the sample was processed to create a HiFi SMRT library, speedy sequencing of four SMRT Cells 8M was performed on the Sequel II System at Radboud UMC. The result was more than two terabytes of raw polymerase data, with an average yield of more than 500 Gb per SMRT Cell.
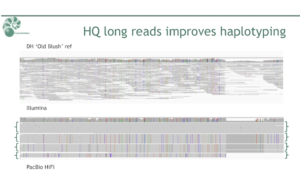
“We did a k-mer analysis to investigate the heterozygosity of the sample. Due to the high accuracy of the reads, we could nicely see four distinct peaks, which you would expect in a heterozygous, tetraploid sample,” Nijland said. “And when mapping the HiFi reads, we could already distinguish four haplotypes. So we were very happy to see this.”
In order to get an even better picture of the variation between the diploid and tetraploid varieties, Nijland and colleagues, including Henri van de Geest (@geesthc) and Mark de Heer, performed a de novo assembly using FALCON and Canu.
“Our assembly is very much improved and we were able to separate many of the haplotypes,” Nijland said.
“We managed to assemble a heterozygous, polyploid genome, without the need for ultra high molecular weight DNA, which is required for a lot of other long-read sequencing,” Nijland said. “Also, the sequence coverage which is required in the assembly is lower, and because of the high accuracy, the computation of the assemblies is much less.”
“Most importantly, we’re getting a better representation and better overview of genomic content in the assembly. This provides a very valuable tool for molecular breeding efforts in rose.”
Catch up on other PAG presentations in a recent blog post and watch Nijland’s full PAG talk here: