Scientists at Stanford University and the Icahn School of Medicine at Mount Sinai have made impressive strides in resolving variants in the SLC6A4 promoter associated with susceptibility to psychiatric disorders and response to antidepressants. This progress was made possible with highly accurate, long-read sequencing, known as HiFi sequencing.
Published in the journal Genes, the paper comes from lead author Mariana Botton, senior author Stuart Scott, and collaborators. It describes a SMRT Sequencing-based approach to analyzing amplicons of the SLC6A4 promoter region, which is noted for “a variable number of homologous 20–24 bp repeats,” the authors write, as well as long, extra-long, short, and extra-short alleles with differing expression. The gene itself is important for pharmacodynamics of antidepressants, one of the most frequently prescribed class of drugs.
As Botton et al. note, identifying key variants within the promoter region is most valuable in the context of haplotypes showing whether variants share an allele. Unfortunately, that information is not easy to access. “Short-read sequencing is not effective at accurately interrogating the SLC6A4 promoter, particularly across the VNTR that includes the 5-HTTLPR insertion/deletion (L>S) polymorphism,” the scientists report. “This overarching limitation of short-read sequencing has previously been acknowledged, as low complexity regions and tandem repeats in the human genome are notoriously challenging for short-read platforms.”
With that in mind, the team turned to long-read sequencing data from PacBio. They designed four overlapping primer sets to span the SLC6A4 promoter region and the necessary oligo tags for barcoding, and then sequenced the resulting amplicons for 120 samples with SMRT Sequencing. They also performed Sanger sequencing and gathered publicly available short-read data for many of the samples and compared genotype results across platforms.
The scientists found that three of the key variants “were either not detectable or incorrectly genotyped among the [various short-read data sets] in 32/32 (100%), 60/68 (88%) and 17/21 (81%) samples and 87/96 (91%), 85/204 (42%) and 34/63 (54%) variant sites, respectively.” PacBio sequencing, on the other hand, allowed for the detection of all variants, including rare extra-long alleles. “In addition to being more accurate at this locus than short-read sequencing, long-read SMRT sequencing also unambiguously phased the polymorphic SLC6A4 promoter in all samples, including complex compound heterozygous diplotypes,” the team adds. Sanger sequencing results for six samples confirmed the variants identified by SMRT Sequencing.
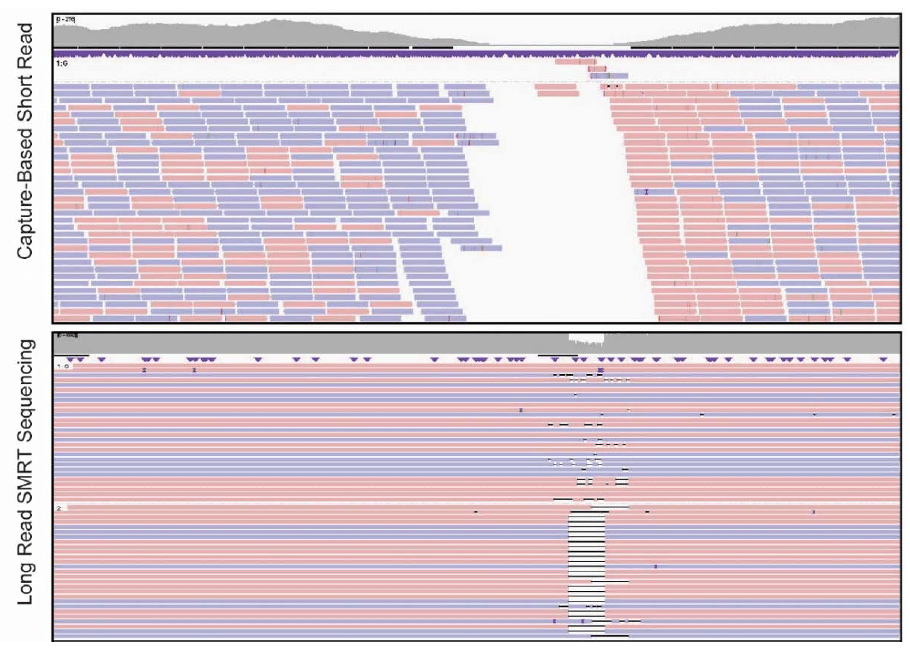
To assess the reproducibility of the SMRT Sequencing workflow, the team evaluated reference samples with known SLC6A4 variants in triplicate. “The intra- and inter-run genotype and diplotype concordances for the 15 control samples were both 100%,” the researchers write.
“Our innovative method enabled the phased resolution of complex SLC6A4 promoter diplotypes, which was not possible using short-read WGS data (~5X and ~45X) or high-depth capture-based short-read sequencing data (~330X),” the team notes. “SLC6A4 long-read SMRT sequencing is a reliable and validated third-generation sequencing technique that can accurately interrogate the low-complexity homologous SLC6A4 promoter region.”
Learn more about best practices and workflows for targeted sequencing in human biomedical research.
December 16, 2020 | Pharmacogenomics