
Rice was the first crop genome ever completed almost two decades ago. However, the rice reference has never been truly complete. Even improved versions of the major food staple and breeding model system Oryza sativa have contained gaps and missing sequences.
An international team of scientists from China, the United States and Saudi Arabia, has finally closed those gaps to produce two gap-free reference genome sequences of the elite O. sativa xian/indica rice varieties Zhenshan 97 (ZS97) and Minghui 63 (MH63).
How Long-Read Sequencing Fills the Gaps
As reported in Molecular Plant, Jianwei Zhang (Huazhong Agricultural University, Wuhan), Jesse Poland (Kansas State University) Rod Wing (Arizona Genomics Institute and KAUST) et al, were able to drill down to centromere level, discovering more than 395 non-TE genes located in centromere regions, of which ~41% are actively transcribed.
Previous references released in 2016 saw 10% of the genome still unassembled/unplaced, and an update in 2018 left eight and seven gaps in the ZS97 and MH63 genomes, respectively.
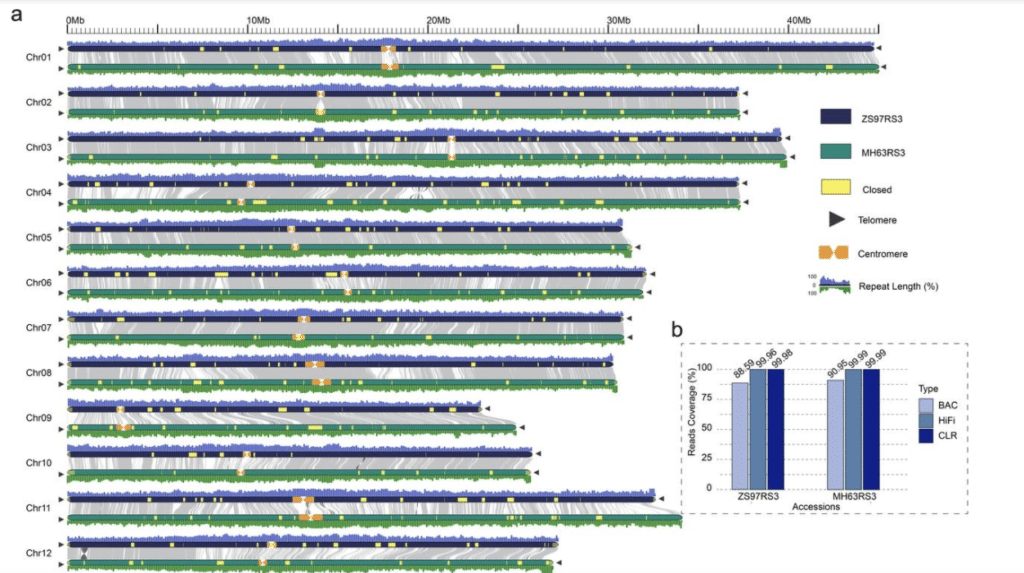
“To bridge all remaining assembly gaps across each genome, we incorporated high-coverage and accurate long-read sequence data and multiple assembly strategies,” the authors wrote. These strategies included both CLR and HiFi sequencing modes.
Hi-C and Bionano maps were used to validate the quality of the assemblies, and FISH and ChIP-Seq assays were utilized to discover and characterize the location and primary structure of centromeres.
The new assemblies captured a 99.88% BUSCO score and LTR assembly index (LAI) numbers that meet the standard of gold/platinum reference genomes. In addition, more than 1,500 rRNAs were identified, compared to tens in the original assemblies.
The last closed gaps in the assemblies were all in centromere regions. Centromeric regions, while critical for fidelity and segregation of chromosomes, are largely inaccessible to breeding due to greatly reduced recombination, particularly in larger genomes, the authors noted.
“The detailed understanding of centromere architecture and gene content, therefore, affords insight into the challenge of developing favorable allele combinations in the absence of natural recombination, using hybrid complementation, gene editing, or even precisely inducing recombination,” they wrote.
With its high accuracy and repeat-spanning reads, PacBio HiFi long-read sequencing was a “great resource for the assembly of complex heterozygous regions and centromeres,” the authors stated.
What the Rice Reference Genome Means for the Future
The large 10-fold variation in the number and distribution of centromeric repeats across the different chromosomes and between the genomes gives a detailed picture of the large amount of centromeric diversity both within and among plant genomes.
The new references provide a clear picture of the primary sequence architecture of the xian/indica rice genomes that feed the world, and could help in the breeding of climate resilient varieties, the authors concluded.
“Such resources will serve to develop a fundamental and comprehensive model for the study of heterosis, and other basic and applied research, and leads the path forward to a new standard for reference genomes in plant biology,” they wrote.
Interested in reading more about long-read sequencing and HiFi reads? Check out our Plant and Animal page to learn more about how they empower insect biology, crop improvements, animal health and breeding and more.
June 25, 2021 | Agrigenomics