What is a pangenome?
Unless you have an identical twin, no other person has a genome that is identical to yours. The same is true for other animal, plant, and microbial species that reproduce sexually: the genomes of individuals are unique. Less well known, but equally true, is that individual members of a species do not always share even the exact same genes. Nevertheless, scientists mostly use a single reference genome to represent an entire species: one human genome, one maize genome, one Staphylococcus aureus genome.
The coining of the “pangenome”
Around 2005, geneticists started to explore the concept of the pangenome, originally defined as the entire set of genes possessed by all members of a particular species and then extended to refer to a collection of all the DNA sequences that occur in a species.
It started with bacteria, as many things do. Genomic activity like recombination, mobile genetic elements, and horizontal gene transfer were clearly contributing to individual diversity across the bacterial domain. Some scientists discovered dozens, if not hundreds, of unknown genes when they sequenced new strains.
In 2007, MIT microbiologist Sallie Chisholm (@ChisholmLab_MIT) set out to determine the extent of genetic variation in the marine cyanobacterium Prochlorococcus. Each strain contains approximately 2,000 genes, and Chisholm estimated that a pangenome for Prochlorococcus would be around 6,000 genes, based on an initial set of 12 genome sequences. Eight years later, with 45 strains sequenced, she revised that estimate up to at least 80,000 genes — around four times the number of genes in the human genome — with the core genome for the species comprising only about 1,000 genes, or less than 2 percent of the total gene pool.
“That’s a lot of information shaping that collective,” Chisholm told The Scientist. “[The pangenome view] changes the way you think about what an organism is.”
Why is it important to capture the full range of genetic diversity?
Those looking to create vaccines need to understand the genomic variation and versatility of disease-causing microbes, especially if they are hoping to develop universal vaccines that could provide protection against more than one strain in a species.
Those studying adaptation to climate change would benefit from a comparison of genes absent or in abundance within species found in different geographic locations and/or environmental conditions. In crop plants, differences in variable genes could have implications on disease resistance, metabolite production, and stress responses.
And with differences in gene number increasingly being associated with disorders including autism, Parkinson’s, and Alzheimer’s diseases, there are strong medical justifications for taking a more variation-centric view of the human species. Variants cannot be identified within regions completely missing from the reference sequence, many of which have been found to be more common than previously thought.
What is being done to generate pangenomes?
To answer this question, we sat down with a few scientists to talk about the era of the pangenome and what’s to come.
One particularly important crop that has haunted geneticists and breeders for years is maize. It is challenging to sequence because the vast majority of its 2.3 Gb genome, a staggering 85 percent, is made up of highly repetitive transposable elements. Maize is also incredibly diverse in its DNA makeup. As an example, a study comparing genome segments from two inbred lines revealed that half of the sequence and one-third of the gene content was not shared — that’s much more diversity within the species than between humans and chimpanzees, which exhibit around 94 percent sequence similarity.
“The whole notion of a single reference genome for crop plants is an antiquated concept borne out of necessity from the technological limitations of the past. Now with the capability to rapidly generate high-quality references for even the largest crop genomes, we can readily access the full complement of sequence diversity and structural variation within a crop,” says Kevin Fengler, Comparative Genomics Lead at Corteva Agriscience.
So the field was delighted when a collective of 33 scientists released a 26-line maize pangenome reference collection earlier this year. The collection was created using PacBio sequencing, and includes comprehensive, high-quality assemblies of 26 inbreds known as the NAM founder lines. These include the most extensively researched maize lines that represent a broad cross section of modern maize diversity, as well as an additional line containing an abnormal chromosome 10.
It turns out it’s not just maize biology that can be informed by pangenomes. “The high level of diversity in maize is well known, but we see a lot of diversity and structural variation underlying traits of interest in all the crop plants we work on. Creating the first reference genome for a crop genome is a great first step, but things get really interesting as you begin to add more genomes and a more comprehensive view emerges,” adds Fengler.
As for our own species, the current reference genome (GRCh38) — an update of the genome produced by the international Human Genome Project in 2000 and based mostly on DNA from one person — has been added to and annotated through the years, but is still an incomplete sequence and woefully inadequate as a representation of human diversity and genetic variation. Scientists estimate that up to 40 megabases of sequence, including protein-coding regions, are absent from the reference genome.
Several studies using PacBio long reads have reported an average of ~20,000 structural variants (SV) per human genome, most of which fall within repetitive elements and segmental duplications. Furthermore, it does not represent the diploid structure of human genomes. Rather, it is an arbitrary linear combination of different haplotypes, or a mosaic of multiple individuals.
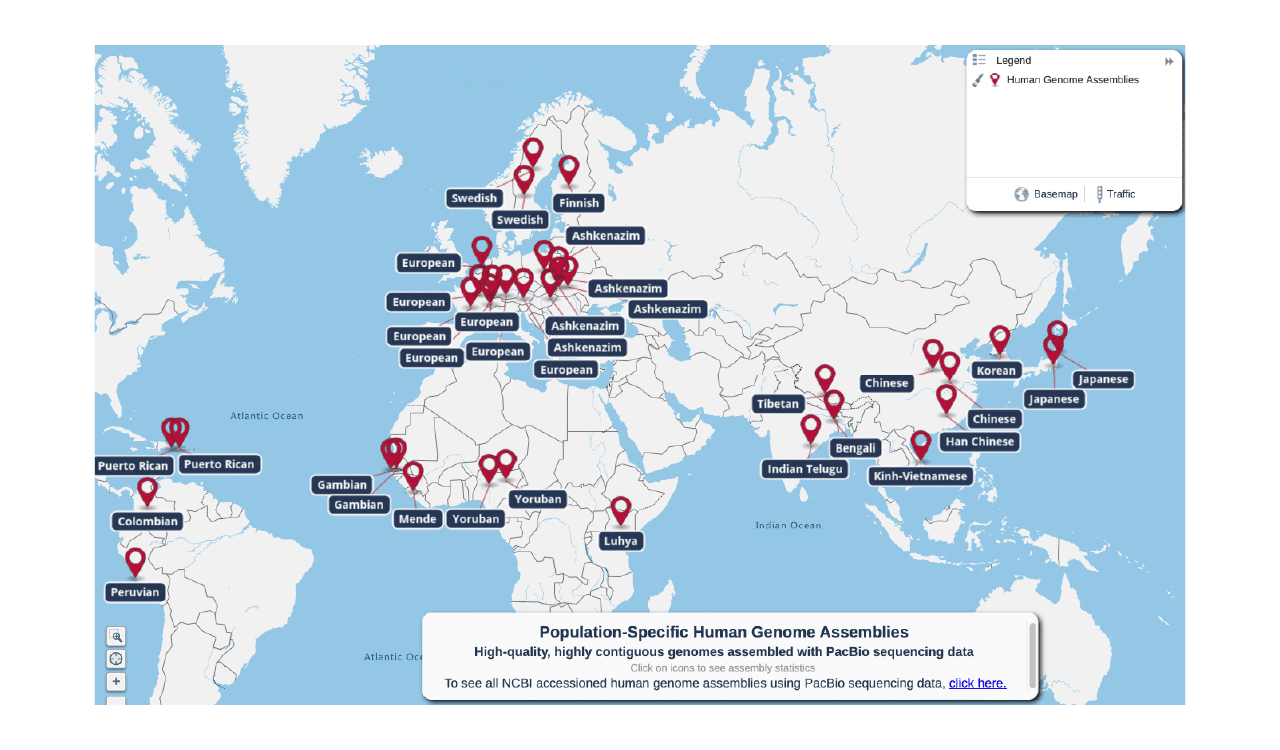
Several groups have undertaken efforts to ensure certain populations are better represented in genomic databases, from Sweden to Tibet to Japan.
When asked the value a pangenome could bring to human research, Fritz Sedlazeck (@sedlazeck), Assistant Professor at Baylor College of Medicine, said, “the pangenome has the potential to represent the diversity of the human population or any species. This eases the re-identification of complex alleles or even haplotypes.”
And it seems the National Human Genome Research Institute agrees, recently committing $30 million towards the creation of a new human pangenome based on high-quality sequencing of 350 individuals from across the human population, to capture all genomic variation observed in human populations.
“One human genome cannot represent all of humanity. The human pangenome reference will be a key step forward for biomedical research and personalized medicine. Not only will we have 350 genomes representing human diversity, they will be vastly higher quality than previous genome sequences,” said David Haussler, director of the University of California Santa Cruz Genomics Institute, which is leading the project.
How to generate a pangenome
So, what are the most important things to keep in mind when creating a pangenome reference?
First, Fengler says that being able to be confident in your results is really important. “Ideally, all of the references in the pangenome collection will be built with a similar recipe to enable direct comparisons without artifacts from different technologies.” This points to the need for a reliable technology that can be used to generate equivalent quality genomes for many samples with little variability.
Second, the data must be high quality. When asked the importance of long reads to pangenome efforts, Sedlazeck said, “they will be important to distinguish between different alleles/paths in the graph and to characterize novel mutations. Thus, being able to cope with graphs that encode a much higher number of variations to better represent the population.” Along those lines, Fengler adds, “the approach for assembly needs to be robust and accurate such that mis-assembly and sequence errors are not interpreted as structural variation and sequence diversity.”
Lastly, cost and speed have to be taken into account. With the high accuracy of HiFi sequencing, only 10- to 15-fold coverage per haplotype is needed for a high-quality resulting genome assembly, and the analysis time can be cut in half.
“Now researchers no longer need to wait for actionable sequence data,” says Fengler. “For maize, we can generate a high-quality reference genome the same day that the sequencing finishes.”
What’s next in pangenomes?
As pangenome collections grow, scientists have to tackle questions around how to represent a pangenome. “Which variations should be included into a pangenome? Is it all of them? Then you lose specificity in regions. Is it only the common variations? Then you have a problem with disease-causing variations and other complex regions like HLA,” asks Sedlazeck, highlighting the continued work that needs to be done.
In addition, tackling things like annotation, visualization, and relationship management are on Fengler’s mind. “A variety of new pangenome analysis and visualization tools are needed to fully realize the value of having a pangenome collection for each crop.”
And then we have to move into functional and translational analysis. Scientists need to be able to take their newfound understanding of variation at the genome level and see how it impacts phenotypes, and whether the variation can be introduced artificially to influence agronomic traits, for instance.
One thing is for sure: the pangenome era is upon us, and whether you need a pangenome to understand important traits or you build tools to interpret those traits, there will be plenty to work on in the coming years.
Interested in finding out more about HiFi data for pangenome sequencing your organism of interest? Get in touch with a PacBio scientist to scope out your project.
Explore other posts in the Sequencing 101 series:
The evolution of DNA sequencing tools
Introduction to PacBio sequencing and the Sequel II system
From DNA to discovery — the steps of SMRT sequencing
Why are long reads important for studying viral genomes?
Understanding accuracy in DNA sequencing
The value of sequencing full-length RNA transcripts of DNA transcripts
Ploidy, haplotypes, and phasing — how to get more from your sequencing data
Video: Sequencing 101: how long-read sequencing improves access to genetic information