 An exciting new paper from scientists at the National Institute of Allergy and Infectious Diseases and the NIH Clinical Center reports on the evolution of the SARS-CoV-2 virus within individuals. The team used HiFi sequencing to make this work possible.
An exciting new paper from scientists at the National Institute of Allergy and Infectious Diseases and the NIH Clinical Center reports on the evolution of the SARS-CoV-2 virus within individuals. The team used HiFi sequencing to make this work possible.
The paper, which was published in PLoS Pathogens, comes from lead authors Sung Hee Ko, Elham Bayat Mokhtari, Prakriti Mudvari, senior author Eli Boritz, and collaborators. They conceived the project to overcome a key challenge in tracking viral adaptation. “An important obstacle to understanding intra-individual evolution of SARS-CoV-2 is that standard sequencing and analytical procedures yield a single consensus sequence for each sample, rather than multiple sequences representing virus quasispecies diversity,” they write.
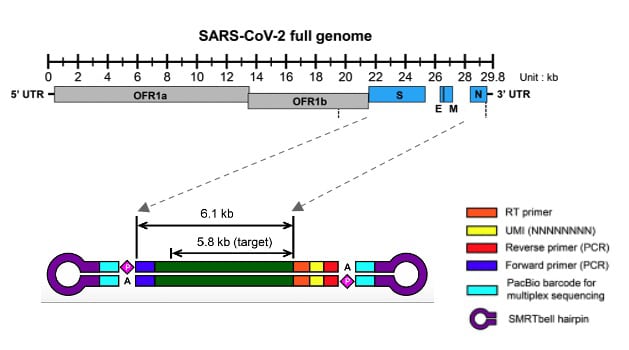
To address the issue, they developed a new method based on HiFi sequencing to focus on the 6.1 kb region of the SARS-CoV-2 genome encoding its surface proteins. They then conducted deep sequencing of eight individuals, yielding large numbers of fully phased S, E, and M gene sequences from each person. In one individual, the availability of four samples collected over time allowed for a longitudinal analysis of viral response to host immune pressure. The scientists had previously used HiFi sequencing to study the intra-individual evolution of HIV, and believed that the same approach could be useful during the COVID-19 pandemic.
The choice of HiFi sequencing, which builds a highly accurate sequence based on consensus calls from covering the same molecule over and over, gave the team an excellent view of viral evolution. When we asked senior author Eli Boritz about his choice of technology, he shared that “By early 2020, we had been working for several years to use HiFi sequencing for high-throughput, single-copy, long-read HIV genetic analysis. Our approach in the HIV studies used unique molecular identifiers (UMIs) for error correction and drew on a short-read approach from Ron Swanstrom’s group and a PacBio approach from Jim Mullins’s group. As the pandemic took off around the world, we decided to adapt our approach to SARS-CoV-2. We didn’t know if this new virus would generate enough diversity to warrant our detailed sequence analysis, but we decided that it would be important to look.”
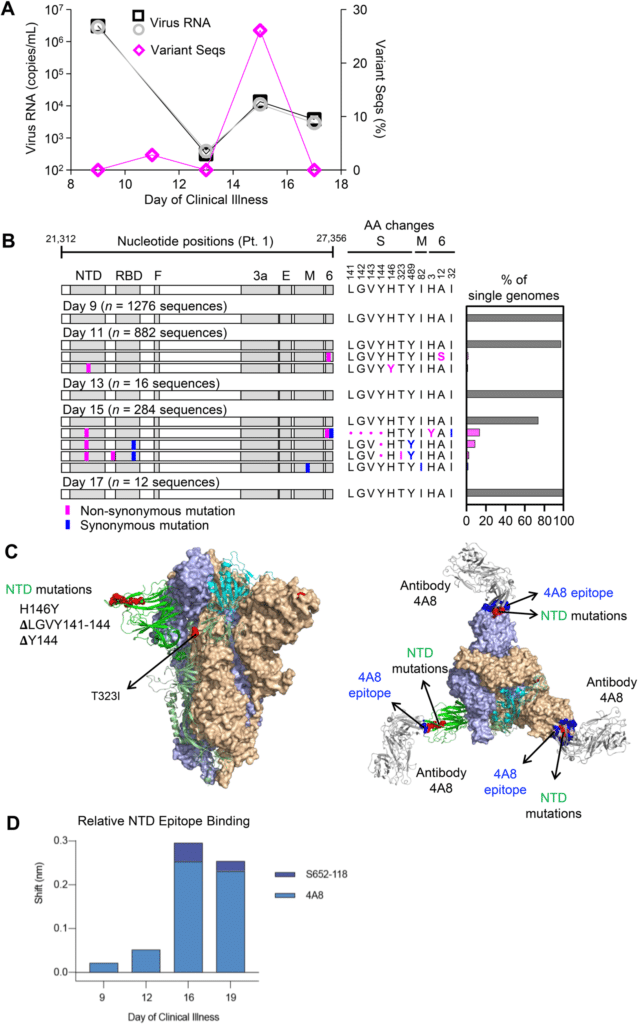
The longitudinal analysis yielded results highly suggestive of natural selection, revealing four viral haplotypes harboring three mutations that arose independently in a single epitope. “These mutations arose coincident with a 6.2-fold rise in serum binding to spike and a transient increase in virus burden,” the scientists note. “We conclude that SARS-CoV-2 exhibits a capacity for rapid genetic adaptation that becomes detectable in vivo with the onset of humoral immunity, with the potential to contribute to delayed virologic clearance in the acute setting.”
In the other study participants for whom repeated sampling was not possible, the team found lower genetic diversity in the viruses sequenced. They hypothesize that this is likely the result of analyzing samples collected early in the infection process rather than after the host’s immune response has had time to select variants with mutated spike proteins.
We asked Eli Boritz about what’s next for his team. For future longitudinal studies, he told us, “it will be important … to sequence additional regions of the virus and to perform a comprehensive analysis of antiviral host responses, including neutralizing antibodies, T cells, and other mechanisms.” He also hopes to analyze viral samples from more complex cases, such as reinfections. “We hope these studies can teach us about the virus’s capacity for additional waves of escape variants in the future,” he said.
The team’s insights into viral evolution in a single person have important implications for COVID-19 treatment. “Our results also emphasize that early antiviral therapy or combinations of antivirals with distinct targets could have markedly higher virologic efficacy than monotherapy administered later in the disease course,” the scientists conclude.
Interested in hearing more about COVID-19 from Dr. Boritz? Access the webinar here.
Or, visit our Coronavirus Sequencing page to learn how HiFi reads from the Sequel IIe System provide a reliable, cost effective, high-throughput solution for sequencing SARS-CoV-2 quickly and easily.