 Tackling larger and larger genomes has been an attractive pursuit for many scientists as sequencing technologies improve at rapid rates. But what about the other end of the spectrum — the tiny organisms that comprise much of the diversity of life?
Tackling larger and larger genomes has been an attractive pursuit for many scientists as sequencing technologies improve at rapid rates. But what about the other end of the spectrum — the tiny organisms that comprise much of the diversity of life?
An obvious obstacle to decoding the DNA of small organisms such as insects, nematodes and other arthropods is collecting enough of it to actually sequence (usually multiple micrograms worth). Until recently, the solution was to pool DNA from many of these tiny creatures to create a representative sample, and extrapolate the biology of the individual constituents from there.
But this is far from ideal, as it does not reflect the true genome of an individual within the species, especially between specimens from the wild and those raised (often through in-breeding) in the lab. Nor does it resolve haplotypes. Plus, it can take a long time to breed and collect enough specimens.
Recently, PacBio announced a way to create high-quality de novo genome assemblies from just a couple hundred nanograms of starting genomic DNA. Gap-less, mega-base scale contiguity of these assemblies could offer many advantages, such as providing insights into promoters, enhancers, repeat elements, large-scale structural variation relative to other species, and many other aspects relative to functional and comparative genomics.
This ‘low DNA input’ workflow was embraced by many, and the first usage, a collaboration with scientists at the UK’s Wellcome Sanger Institute, resulted in the assembly of an Anopheles coluzzii mosquito genome with unamplified DNA from a single individual female insect.
Erin Bernberg (@ErinBernberg) of the University of Delaware Sequencing and Genotyping Center, a PacBio certified service provider, took sequencing to another extreme when she extracted DNA from 1.5 cm ice worms and successfully helped Scott Hotaling (@MtnScience) at Washington State University delve into the genomics of the annelids.
“Low-input works,” Bernberg said during a webinar about the work. “You don’t need as much DNA as everyone is worrying about with PacBio. You can generate good data with it.”
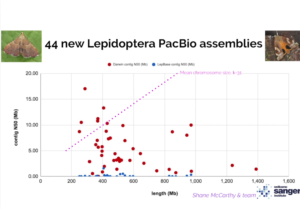
Since then, the workflow has been adapted to the high-throughput Sequel II System and adopted by the Darwin Tree of Life project, which aims to sequence all 60,000 species in the UK, many of which are too small for standard long-read sequencing. Mark Blaxter, Tree of Life Genomics Programme Lead, reported 44 new high-quality Lepidoptera genomes completed with the low DNA input workflow at the Plant and Animal Genomes Conference in early 2020.
And Sanger scientist Chris Laumer (@tendersombrero) shared his work on optimised protocols for sequencing individual meiofaunal organisms using long-range PCR library amplification, HiFi sequencing, and linked-read scaffolding, in a SMRT Leiden presentation.
How low can you go?

Scientists at the Max Planck Institute in Germany are testing whether it is possible to reduce DNA sample sizes even further – to as little as 5 ng of genomic DNA.
Franziska Beran, group leader at the MPI for Chemical Ecology, Bruno Huettel, Head of the Max Planck Genome-centre Cologne (MP-GC), and Christian Woehle, bioinformatician at MP-GC, are using an amplification-based ultra low DNA input workflow to investigate the genome of the horseradish flea beetle (Phyllotreta armoraciae).

The tiny beetle has a fascinating defense strategy known as sequestration, in which the beetles not only overcome the pungent chemical compounds of the horseradish plant, but exploit those chemicals to make themselves unpalatable for their own enemies.
The team has managed to extract valuable genomic data from just 5 ng of genomic DNA from single bugs. They also worked with specimens which were maintained in ethanol, a standard long-term specimen storage solution, suggesting that it may now be possible to sequence specimens from historical collections.
“With this new solution from PacBio, we can now touch specimens which were not possible previously to apply long-read sequencing,” Huettel said. “In the past, we could pool several individuals, but this would produce a mess of data which cannot be resolved into single unique contigs, as we would sequence several haplotypes.”
Together with colleagues at the LOEWE Centre for Translational Biodiversity Genomics (TBG) in Senckenberg, Frankfurt, the MP-GC is also using the ultra low DNA input workflow to delve into the genomics of springtails — minute arthropods that are distant relatives of insects.

Present in the oldest known terrestrial ecosystems (~410 Ma) they are now diversified in a wide range of ecological niches, from tree canopies to ice-cold regions and deep cave systems. They are among the most abundant organisms in soil and thus a major component of soil functions.
Scientists are collecting DNA from two types of springtails: Sminthurides aquaticus and Desoria tigrina. The former is a palearctic species living on floating leaves and wet rocks in ponds, and it is one of the few springtail species displaying a strong sexual dimorphism. The latter belongs to a complex of species common in compost, and thus a commensal of agricultural and gardening activities.
The springtails were sequenced as part of the MetaInvert project, which establishes a database of genomes for several hundred soil invertebrate species, including springtails, oribatid mites, nematodes, potworms, myriapods, and several other groups. These genomes are used to reveal evolutionary relationships, special adaptations and host-microbiome associations. Check out their pre-print for more details about this research.
“The gathering of high-quality genomes is a needed resource to understand the evolutionary history and modalities of terrestrialization of arthropods and identifying the genomic traits that allowed the successful diversification of springtails,” said Clément Schneider of the TBG. “Among the applied prospects is the discovery of new natural products and better management of our soils through a better understanding of the functions of their inhabitants.”
Hear more from PacBio experts and users in the recent webinar, No Organism Too Small: Build High-Quality Genome Assemblies of Small Organisms with HiFi Sequencing.
Read more about the low & ultra-low DNA input workflows:
New Low-Input Protocol Enables High-Quality Genome Created from Single Mosquito
Sequencing at the Extremes: Low DNA Input Workflow Enables Study of Tiny Ice Worm with Giant Genome
Procedure & Checklist: Preparing HiFi Libraries from Low DNA Input Using SMRTbell Express Template Prep Kit 2.0
Now Available: Ultra-Low DNA Input Workflow for SMRT Sequencing
Application Note: Considerations for Using the Low and Ultra-Low DNA Input Workflows for Whole Genome Sequencing