It was the coolest critter Erin Bernberg (@ErinBernberg) had ever worked with – quite literally.
The senior scientist at the University of Delaware Sequencing and Genotyping Center, a PacBio certified service provider, received a shipment of tiny, live ice worms from Washington State University and immediately faced several challenges. How would she get them out of their ice cubes? How would she isolate DNA from the delicate, dark pigmented creatures? And would she be able to extract enough DNA to sequence?
Thanks to the new PacBio low DNA input protocol, the answer to the last question was yes. In fact, Bernberg was able to create a library with enough yield for up to 17 SMRT Cells 1M from only 500 ng of DNA.
She detailed her process for extracting ice worm DNA and shared some of the results during a recent webinar about the new low DNA input workflow.
“Low-input works,” Bernberg said. “You don’t need as much DNA as everyone is worrying about with PacBio. You can generate good data with it.”
Tiny body, giant genome

The ice worm may be tiny at ~1.5 cm, but its genome is giant compared to other annelids, around 1.5 Gb.
Previous attempts to sequence the genome failed because it was “inconveniently large” and the DNA came from pools of several specimens, which clouded the assembly with individual-specific noise, according to Scott Hotaling (@mtn_science), a postdoctoral researcher in the lab of Joanna Kelley at Washington State University who is studying the ice worm as part of his research into species that have adapted to the harsh conditions in extreme environments, like polar regions.
Hotaling said he is excited to delve into the 160 Gb of data generated by Bernberg’s 10 SMRT Cells. He said the genome they have assembled is around 1,000-fold more contiguous than that of its closest relative, the earthworm.
Why is contiguity important in the ice worm genome? There are so many unknowns when it comes to annelid genetics, Hotaling said. As an example, he cited AMP deaminase, a known regulator of energy metabolism that is suspected to play a part in the ice worm’s unique method of thermal regulation.
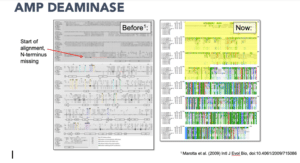
“Ice worms do this cool thing where they actually ramp up their energy levels as they get colder, which is essentially the inverse of what a lot of organisms do,” Hotaling said.
A partial sequence of AMP deaminase (540 amino acids long) had been previously generated for iceworms, so Hotaling was able to easily locate it in his new PacBio generated genome. In doing so, he discovered just how much information had been missing: “tons.”
“Imagine this across 20,000 genes. There’s so much more power to look from gene to gene at variation that might be evolutionarily relevant,” he said.
Evolution at the extremes

Mountain glacier habitats may look desolate, but there’s a lot going on below the surface. Not only are there millions of microbes, but also snow algal blooms and other lifeforms. Among them, ice worms (Mesenchytraeus solifugus) are the largest to spend their life cycle in ice.
The ice worms that Hotaling studies on Mt. Rainier survive extremely harsh conditions. Tens of millions of ice worms can live on a single glacier. Not only do they face constant cold stress, but the highly reflective ice and snow surfaces make the glaciers some of the most UV intensive places in the world.
It’s unclear how they have adopted to such an extreme lifestyle, but such knowledge could provide insight into evolutionary processes, Hotaling said. He wants to learn whether their extreme lifestyle translates to extreme physiology, and how. He will be exploring which genes and pathways have been under selection in ice worm genomes, and whether there is congruence between genes under selection and those differentially expressed in response to stress.
“We want to know things like can they freeze? It seems like a dumb question because they live in ice, but scientific observations from 30 years ago suggest they can only handle temperatures within a few degrees of freezing,” Hotaling said.
Early experiments suggest the worms are living at the absolute lower limit of their thermal tolerance, and they also exhibit a high tolerance for UV. His team is now fleshing out the picture by layering RNA-based annotations on top of the main sequencing data.
“Lineages like the ice worm are pretty far from anything else that’s been sequenced. So just looking at what’s in there is, in its own right, a pretty interesting pursuit. But we’d also like to add some context to look at genome evolution and gene expression,” he said.
He is also keen to taxonomically annotate the gene to capture contamination from other organisms that the ice worms may have hosted.
“We sequenced an entire worm – all of its gut contents, all of its parasites, whatever was on its body. So contamination is almost a certainty,” he said. “I think this is going to be a big challenge for anyone working in this space. How do we develop tools to really deal with this in a high-powered, efficient way?”
For further information on this topic and specifics of the low DNA input workflow, watch the full webinar and read the application note referenced in the presentation. Hear how other scientists are using low and ultra-low DNA input workflows to advance their research.