UPDATED
January 18, 2019
This paper is now available at Genes.
ORIGINAL POST
December 19, 2018
High-quality reference and de novo genomes have been celebrated by geneticists, population biologists and conservationists alike, but it’s been a dream deferred for entomologists and others grappling with limited DNA samples, due to previous relatively high DNA input requirements (~5 μg for standard library protocol).
A new low-input protocol now makes it possible to create high-quality de novo genome assemblies from just 100 ng of starting genomic DNA, without the need for time-consuming inbreeding or pooling strategies. The targeted release date for the protocol is February 2019.
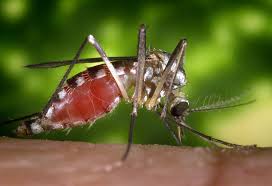
The protocol, developed as a collaboration by scientists at the Wellcome Sanger Institute and PacBio, was used to assemble the genome of an Anopheles coluzzii mosquito with unamplified DNA from a single individual female insect.
As described in a bioXriv pre-print, Sarah B. Kingan, Haynes Heaton, et al. used a modified SMRTbell library construction protocol without DNA shearing and size selection to facilitate the use of lower input amounts, as shearing and clean up steps typically lead to loss of DNA material.
“This new low-input approach puts PacBio-based assemblies in reach for small and highly heterozygous organisms that comprise much of the diversity of life,” said co-corresponding author Jonas Korlach, our chief scientific officer.
The sample was run on the Sequel System with the latest v6.0 software, followed by de novo genome assembly with FALCON-Unzip, resulting in a highly continuous (contig N50 3.5 Mb) and complete (more than 98% of conserved genes were present and full-length) genome assembly.
About a third of the new de novo genome is haplotype-resolved and represented as two separate sequences for the two alleles, providing additional information about the extent and structure of heterozygosity that was not available in previous assemblies, all of which were constructed from many pooled individuals.
“The ability to generate high-quality genomes from single individuals greatly simplifies the assembly process and interpretation, and will allow far clearer lineage and evolutionary conclusions from the sequencing of members of different populations and species,” the authors state.
The first Anopheles gambiae genome, published in 2002, was created using BACs and Sanger sequencing. Further work over the years to order and orient contigs improved this reference and to date, AgamP4 remains the highest quality Anopheles genome among the 21 that have now been sequenced. However, AgamP4 still has 6,302 gaps of Ns in the primary chromosome scaffolds and a large bin of unplaced contigs known as the “UNKN” (unknown) chromosome.
The Sanger/PacBio single-insect assembly was able to place 667 (>90%) of the genes on the UNKN contigs into their appropriate chromosomal contexts.
The assembly’s “gap-less mega-base scale contiguity” will also provide insights into promoters, enhancers, repeat elements, large-scale structural variation relative to other species, and many other aspects relative to functional and comparative genomics questions, the authors state.
The protocol’s potential could also extend to other areas with typically low DNA input regimes, such as metagenomic community characterizations of small biofilms, DNA isolated from needle biopsy samples, and minimization of amplification cycles for targeted or single-cell sequencing applications, the authors add.
Read more about the low & ultra-low DNA input workflows:
Now Available: Low DNA Input Workflow for SMRT Sequencing
Now Available: Ultra-Low DNA Input Workflow for SMRT Sequencing
Don’t Sweat The Small Stuff: Low DNA Input Workflow Enables Sequencing of the Smallest Species
December 19, 2018 | Plant + animal biology