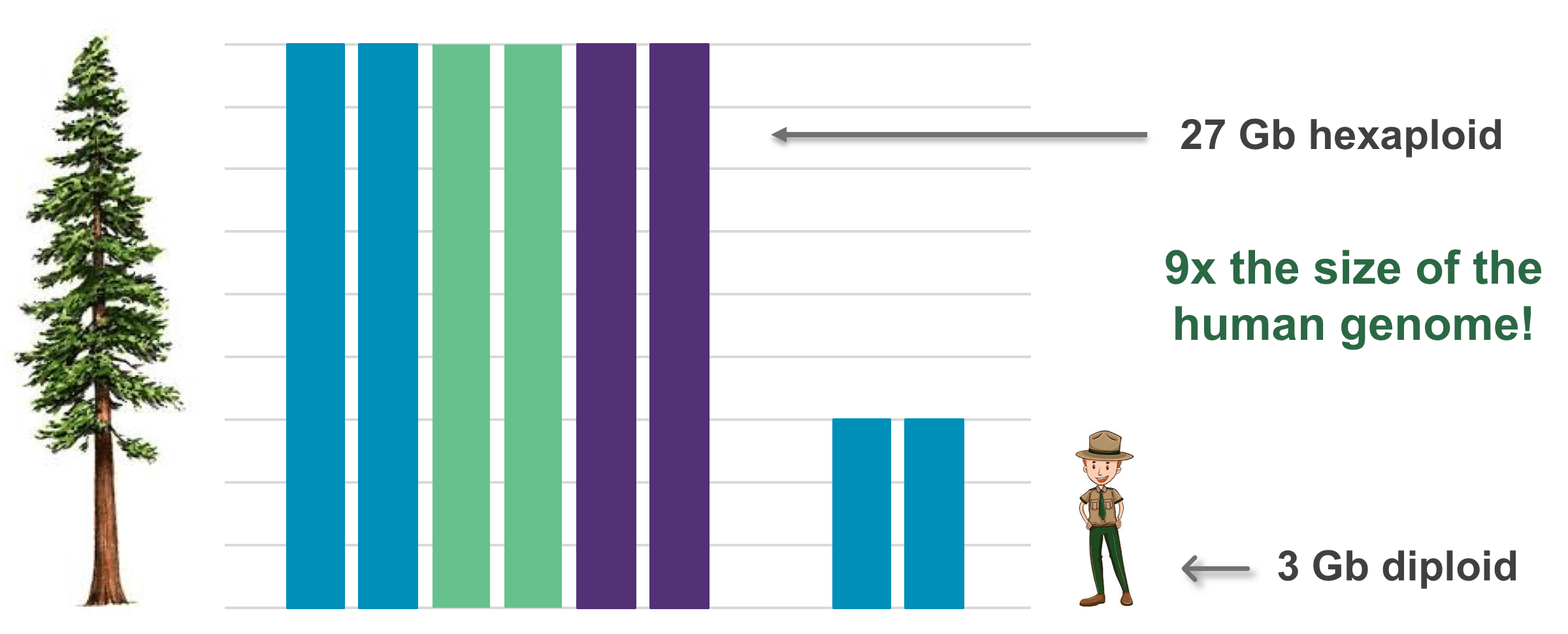
California redwoods: Not only are they giants in height and age (up to 379 feet high, 29 feet round, and thousands of years old), but the famous towering trees are also derived from a massive 27 Gb genome.
Seeking a sequencing challenge for the Sequel II System, we picked the California redwood, or Sequoia sempervirens as it’s known to scientists. There also happened to be several fine specimens at nearby Stanford University.
A small crew of PacBio scientists — Emily Hatas (@EmilyHatas), Greg Young (@PacbioGreg), and Michelle Vierra (@the_mvierra) — headed to campus to acquire samples equipped with ice, scissors, and a kitchen scale. DNA was isolated (using the Circulomics Plant Nuclei kit), a HiFi library was created, and sequencing got underway. In just seven days, the team achieved 22-fold coverage of the genome (606 Gb of HiFi data). Another 6 days later, Greg Concepcion (@phototrophic) generated a partially haplotype-resolved genome assembly almost twice the expected genome size with a contig N50 of 1.92 Mb.
“The results were amazing,” Vierra said. “We are very pleased to see the improvements that this genome assembly represents over other recent conifer genomes.”
 The massive genome was put together in just 17 days — 4 days of sample prep, 7 days of sequencing, and 6 days for assembly, and was detailed in a Medium post by Vierra.
The massive genome was put together in just 17 days — 4 days of sample prep, 7 days of sequencing, and 6 days for assembly, and was detailed in a Medium post by Vierra.
But the team wasn’t done yet. As a general recommendation, 10- to 15-fold coverage in HiFi reads is the ideal range to yield a genome that measures up favorable in the 3 C’s of genome quality.
For genomes of the California redwood’s size, it may not be economical or feasible to obtain that much coverage in a limited timeframe, which is why the team opted to see what a reasonable ~20-fold coverage could generate. However, with a little more time on their hands, and enough HiFi library to go around, the team embarked on more sequencing to bring the total to 875 Gb of HiFi data representing 33-fold coverage of the genome.
Not surprisingly, more of a good thing (HiFi reads) made an even better genome assembly. The contiguity improved significantly with a contig N50 of 3.8 Mb and completeness increased with an almost 61% complete BUSCO score.
Overall, what would have been considered a herculean effort not that many years ago was accomplished in only a few weeks by a handful of personnel in their spare time. It’s our hope that with the increasing adoption of PacBio HiFi reads we will continue to see massive improvements in the assembly of all genomes, including large and increasingly complex polyploid plants.
The additional data for the redwood genome has been made publicly available along with the updated genome assembly and can be found here. We hope it will be a useful tool for conifer researchers everywhere!
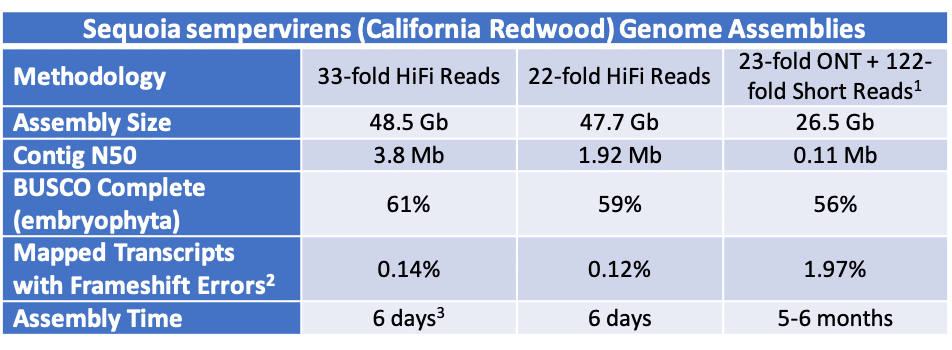
Comparison of California Redwood genome assembly results. [1] Hybrid assembly of redwood. [2] Transcript set of Abies alba from Neale et al. Varying number of transcripts aligned to each genome (4,958 mapped to 22-fold HiFi Reads, 4,970 mapped to 33-fold HiFi reads, 4,760 mapped to ONT) [3] Assembly with 33-fold HiFi reads was done with 80 cores and an updated version of Hifiasm (0.3.0).
—
Interested in finding out more about HiFi data for sequencing your organism of interest? Get in touch with a PacBio scientist to scope out your project.
April 15, 2020 | Plant + animal biology