The scientific community is still abuzz from last week’s announcement of the amazing achievement of the first complete sequence of a human genome by the Telomere-to-Telomere (T2T) Consortium. Assembling genomes from head to toe with high completeness, contiguity and accuracy has now become a reality, thanks to the advances of long-read sequencing technologies. Indeed, fully finished genomes were listed first among the “Technologies to watch in 2022” in Nature’s annual feature of technologies that look to shake up science this year.
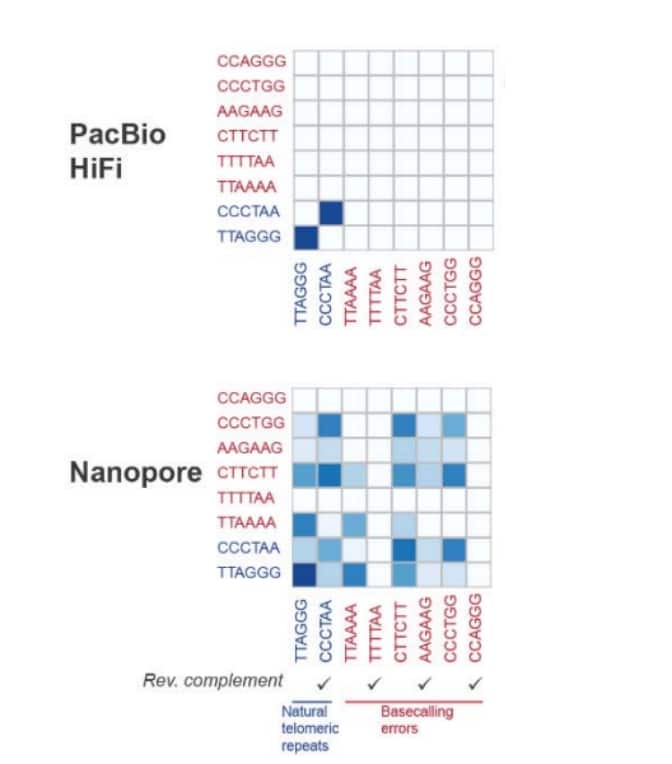
To generate telomere-to-telomere genomes, among other things one needs to be able to sequence is – obviously – telomeres. In a recent preprint posted by Drs. Matthew Meyerson, Heng Li and colleagues (Dana-Farber Cancer Institute, Broad Institute, Harvard University), a comparison of long-read sequencing technologies to sequence telomeres was performed. PacBio HiFi sequencing showed excellent performance, fully resolving the length and sequence content of telomeres using data from the recently sequenced and assembled CHM13 sample of the T2T Consortium. In contrast, Nanopore-based long-read sequencing showed extensive base calling induced errors at telomere repeats.
Similar findings have been reported by others. A recent article in Genome Research by Dr. Chris Mason (Weill Cornell) & collaborators highlights the ability of PacBio’s accurate and long HiFi reads to resolve the two telomere alleles in diploid genomes, and observes new haplotype diversity and sequence heterogeneity in human telomeres.
Beyond human genome sequencing, fully assembled telomeres are now being observed regularly in plant & animal genome HiFi assemblies. In one recent example, a recent preprint by Yu et al. used PacBio sequencing of telomeres to discover the first known occurrence of telomerase-independent resolution of the end-replication problem within chordate animals. The highly regenerative newt Pleurodeles waltl lacks telomerase activity, contains telomeres distinct from all known vertebrates in both sequence and structure, and deploys an alternative lengthening of telomeres (ALT) mechanism for physiological telomere maintenance. ALT has also been observed for the compensation of telomere loss in a subset of human cancer cell lines.
With the importance to produce fully finished genomes, their biological significance in areas such as aging or cancer, the ability to accurately sequence telomeres – and the rest of the genome – accuracy and completeness is paramount. PacBio HiFi sequencing gives researchers confidence in their data to provide the most solid foundation for their research.
Read more about the HiFi Difference
More precise genomes for precision medicine
Not all gigabases are created equal