“We are now embarking on an era where all genetic variation in an individual will be completely discovered,” write Glennis Logsdon (@glennis_logsdon), Mitchell Vollger (@mrvollger), and Evan Eichler in a recent Nature Reviews Genetics paper. “Hundreds and ultimately thousands of new human reference genomes will be produced.” A decade ago that would have sounded impossible, but today this bold proclamation is widely accepted in the genomics community — a telling sign of the remarkable innovation that has driven genome sequencing in recent years.
In their review, the University of Washington scientists give credit for much of these accomplishments to advancements in long-read sequencing. “Sequencing technology is the ‘microscope’ by which geneticists study genetic variation,” they write, “and it is clear that long- read technologies have provided us with a new ‘lens and objective’ for understanding DNA and RNA variation, structure and organization.”
That new lens has allowed researchers to fill in many of the blind spots left by short-read sequencing, which is limited to read lengths of just a few hundred bases. These “are too short to detect more than 70% of human genome structural variation (that is differences that involve 50 bp or more), with intermediate-size structural variation (less than 2 kb) especially under- represented,” the authors note. Long reads generated using PacBio sequencing, on the other hand, can span tens of kilobases.
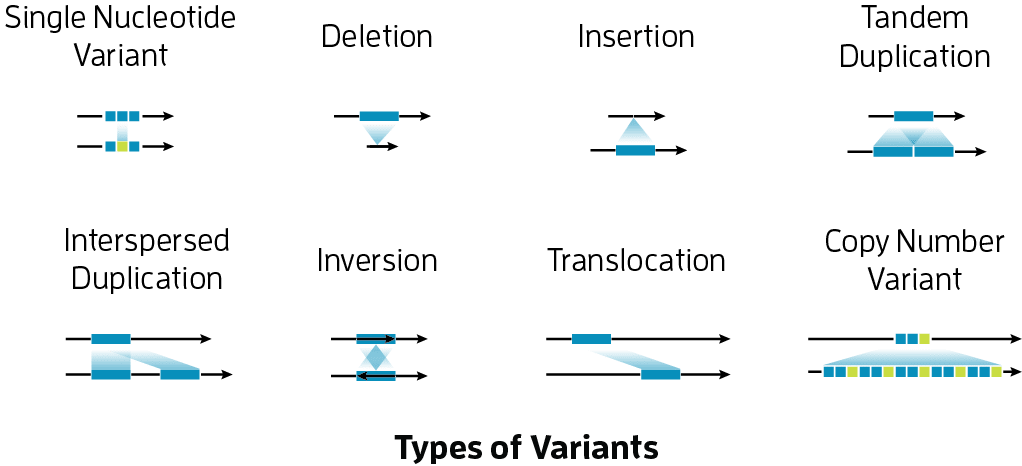

Short-read sequencing platforms also struggle to get through repetitive regions or regions with extreme GC content. “For example, even PCR-free, short-read genomic libraries show up to twofold reductions in sequence coverage when the GC composition exceeds 45%, limiting the ability to discover genetic variation in some of the most functionally important regions of our genome,” the scientists report. Such regions include first exons, centromeres, telomeres, and segmental duplications.
Approaches to extend the capabilities of short reads — such as linked reads, synthetic long reads, and Hi-C — “are generally inferior to strict long-read sequencing approaches” for many applications, the authors write.
The review provides a great education on the applications of long-read sequencing, such as detecting structural variation, enabling diploid and even telomere-to-telomere human assemblies, and characterizing the transcriptome. The authors also explain the various long-read data types, including PacBio HiFi reads, “the first data type that is both long (greater than 10 kb in length) and highly accurate (greater than 99%).” With HiFi reads, the scientists add, it is not necessary to use short-read data for error correction.

The accuracy of HiFi reads, combined with the throughput of the Sequel II System, provide a cost-effective option for variant discovery in population-scale sequencing or family-based sequencing, the scientists note. Even lower 10- to 15-fold HiFi read coverage is useful for finding meaningful variation. With diploid assemblies, long-read sequencing “will revolutionize genomics by revealing the full spectrum of human genetic variation, resolving some of the missing heritability and leading to the discovery of novel mechanisms of disease,” Logsdon et al. write.
“The wealth of additional information afforded by single-molecule, long-read sequencing compared with short-read sequencing promises a more comprehensive understanding of genetic, epigenetic and transcriptomic variation and its relationship to human phenotype,” the scientists conclude.
Explore workflows and additional resources on comprehensive variant detection or structural variant detection.
July 21, 2020 | Human genetics research