For the thousands of scientists who attended The Plant and Animal Genome Conference in San Diego this January, the sentiment seemed to be “ask not if PacBio is for you, but how PacBio can work best for you.”
The answer that emerged during PacBio’s PAG workshop and subsequent SMRT Informatics Developers Conference was a complex one.
Recent developments, such as new chemistry, new SMRT Cells, the SMRTbell Express Template Prep Kit, and SMRT Link 6.0 software have already led to faster and easier library prep, longer reads with more data and reliability, better transcript characterization (Iso-Seq) and phasing (FALCON-Unzip) capabilities (discussed by PacBio principal scientist Liz Tseng), and deeper insight with less waiting.
As senior product manager Justin Blethrow laid out in his talk, “Sequence with Confidence – How SMRT Sequencing is Accelerating Plant and Animal Genomics,” the upcoming release of a new Express Template Prep Kit will make the faster library prep available for more applications, and the Sequel II System will provide 8-times the amount of data while also integrating circular consensus sequencing. Also in the product roadmap for 2019 is a low DNA input protocol for limited samples and the smallest of organisms.
Big data from tiny samples
A talk at the PacBio PAG workshop by Andrew Clark of Cornell University gave attendees a sneak peek at one of these exciting opportunities. In a collaboration with Manyuan Long of the University of Chicago and Rod Wing of the University of Arizona, the evolutionary ecologist was able to use PacBio sequencing to create new genome assemblies of 10 drosophila species, including de novo assemblies of two individual flies, using as little as 26 ng of gDNA. Clark was most curious about why D. virilis diverges so dramatically from other species in terms of heterochromatic regions with long sections of simple satellite repeats — up to 40% of the genome.
“These regions of the genomes are really a bear to work with,” he said. But with PacBio long-read sequencing, the assemblies “flew together nicely,” with almost whole chromosome arms assembling into single contigs. And patterns were readily apparent after taking a look at the raw reads.
“This method to develop whole genome sequencing from single individuals is terrifically exciting in terms of the kinds of new questions that we can generate and answer with those data,” Clark added.
The pangenome era
Other speakers at the PAG workshop heralded the dawn of the pangenome era and delved into detail about their work to create multiple references for plant and animal species.
Max Planck researcher Sonja Vernes, director of the Bat1K consortium, discussed her group’s ambitious efforts to sequence the genome of every living bat species.
Initial data shows clear improvement in the quality of assemblies generated with long reads, she said. The PacBio assembly of the greater horseshoe bat (Rhinolophus ferrumequinum), for instance, contained just 679 contigs, to a standard of 19.9 Mbp NG50, compared to its previously posted assembly made up of 290,000 contigs at a standard of 0.01 Mbp NG50.
Isoform sequence (Iso-Seq) analysis has also provided a wealth of information about transcripts that differ between different sites throughout the body, enabling comprehensive genome annotation.
“We’re really missing out on this information if we don’t go in and collect the functional data to understand the gene structure,” Vernes said.
Kevin Fengler, of Corteva Agriscience, described his work with maize. As he pointed out, genome assemblies must be very accurate and robust to be research-ready, which is why he favors a combination of the latest PacBio technology and old-fashioned manual curation to elevate scaffolds to platinum-grade assemblies.
“Base pair error is not sequence diversity, and mis-assembly is not structural diversity,” Fengler said.
He described his workflow and scaffolding assembly across maize pangenome lines, then moved on to the bigger question: what now?
“Here’s really where the fun begins,” Fengler said as he went on to present some pangenome visualization tools, including TagDots, “rapid dot blots for the pangenome era,” and PANDA (PANgenome Diversity Alignments).
A magical world: The Sequel’s sequel
 At SMRT Informatics, Jeremy Schmutz of the HudsonAlpha Institute for Biotechnology and the DOE Joint Genome Institute, charted the evolution of plant sequencing over the last 10 years and gave a glimpse at its future using the Sequel II System.
At SMRT Informatics, Jeremy Schmutz of the HudsonAlpha Institute for Biotechnology and the DOE Joint Genome Institute, charted the evolution of plant sequencing over the last 10 years and gave a glimpse at its future using the Sequel II System.
A 2008 iteration of the soybean genome done with 15M Sanger reads cost 200 times the amount needed to create a more complete genome using 13 Sequel SMRT Cells in 2018, Schmutz said.
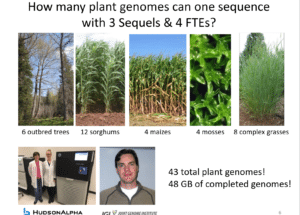
He provided more fun facts. Just how many plant genomes can one sequence with three Sequels and four full-time equivalent staff? Forty-three, Schmutz said — 6 outbred trees, 12 sorghums, 4 maizes, 4 mosses and 8 complex grasses — for a total of 48 Gb of completed genomes.
And his preview of the latest advances included a project to catalog somatic genetic and epigenetic mutations across 200-year-old poplar trees. Circular consensus sequencing (CCS) to generate HiFi reads on the new Sequel II allowed scientists to capture and correlate data from several sites along the trees, such as individual branches, as well as call SNPs, detect structural variants, and phase haplotypes.
“What can we do with the new Sequel IIs?” he asked the crowd. “Tackle outrageous sized plants, create high-quality pangenomes of species, use CCS for metagenomes, precise SNP and structural variation detection, phase haplotypes for alignable regions, and develop new hybrid, outbred, polyploid strategies.”
Other speakers at the half-day event also heralded the new HiFi paradigm, and the final session turned into friendly debates — about the ideal default parameter set for Iso-Seq to get the best bang for your buck, between PacBio scientist Liz Tseng and Roslin Institute bioinformatician Richard Kuo; and the speed of the CCS algorithm (used to generate HiFi data) and its feasibility in large-scale studies, between PacBio algorithm expert Jim Drake, HudsonAlpha’s Jeremy Schmutz and Sergey Koren of the National Human Genome Research Institute.
The next SMRT Scientific Symposium and Informatics Developers Meeting will take place May 7 – 9 in Leiden, Netherlands. Registration is now open.
PacBio posters from PAG can be viewed here:
- “Library Prep and Bioinformatics Improvements for Full-Length Transcript Sequencing on the PacBio Sequel System” – Michelle Vierra, et al
- “A Low DNA Input Protocol for High-quality PacBio De Novo Genome Assemblies from Single Invertebrate Individuals” – Sarah B. Kingan, et al.
- “Haplotyping Using Full-Length Transcript Sequencing Reveals Allele-Specific Expression” –
Elizabeth Tseng, et al. - “Single Molecule High-Fidelity (HiFi) Sequencing with >10 kb Libraries” – Paul Peluso, et al.