
They may not be as well known as our chimpanzee or gorilla cousins, but macaques have played many key roles in scientific progress over the last half century. From launching into orbit during the early days of space travel to revealing the genetics of neurodevelopmental disorders and infectious diseases today, the rhesus macaque remains a key research primate around the world.
A new, comprehensively annotated reference genome unveiled last month boosts the potential of the most widely used non-human primate in biomedical research even further, with new insight into gene functions and disease susceptibility.
A large team of researchers — led by Wesley C. Warren of the University of Missouri; Evan E. Eichler of the University of Washington; and Jeffrey Rogers of Baylor College of Medicine — has released an updated rhesus macaque (Macaca mulatta) reference genome that increases the sequence contiguity 120-fold over previous assemblies.
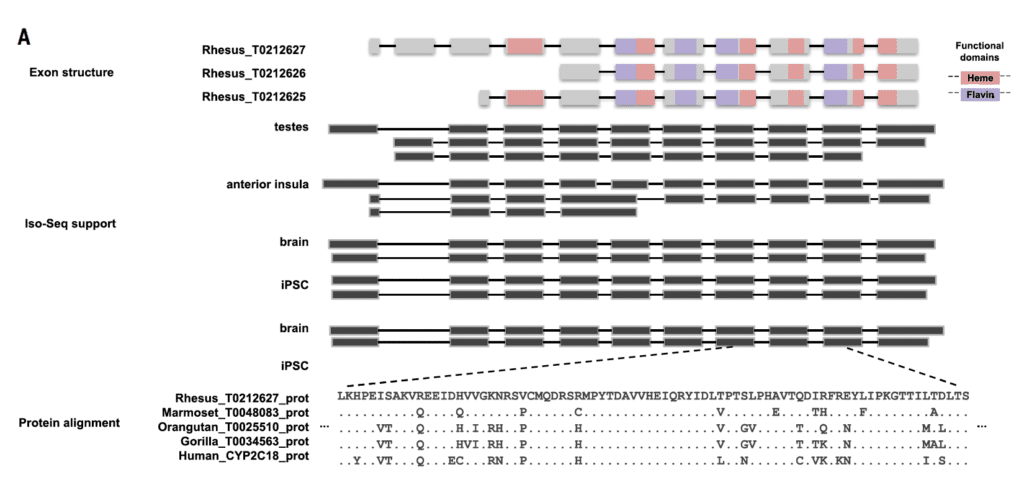
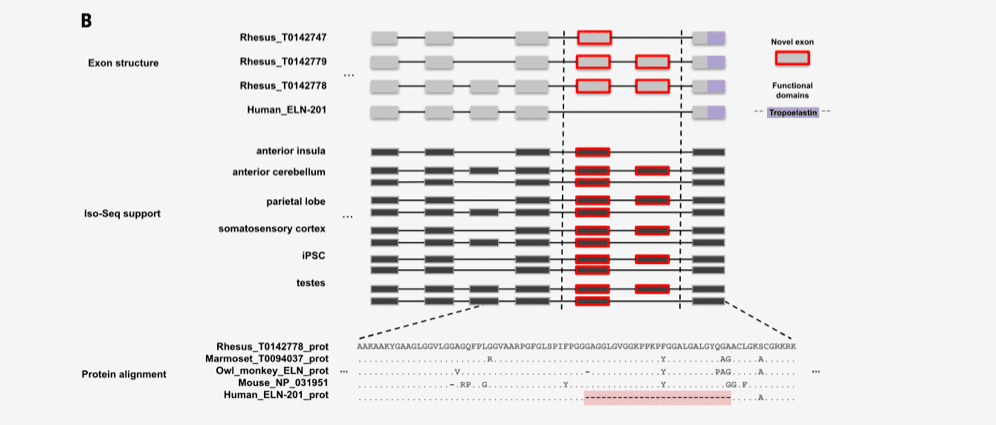
They also used PacBio full-length isoform sequencing, the Iso-Seq method, to analyze 6.5 million full-length transcripts and create a comprehensive set of protein-coding and non-coding gene models. This provided vital information about gene content, organization and isoform diversity, and led to the identification of new macaque isoforms and gene candidates.
“With the improved assembly of segmental duplications, we discovered new lineage-specific genes and expanded gene families that are potentially informative in studies of evolution and disease susceptibility,” the authors wrote.
Bigger picture
In addition to the new reference, the team gathered whole genome sequencing data from 850 rhesus macaques from U.S. research colonies and three wild-caught Chinese samples. Similar to the human 1000 Genomes project, they wanted to catalog genetic variation within the species, and ended up identifying 85.7 million single-nucleotide variants (SNVs) and 10.5 million indels — the most extensive collection of segregating genetic variants for any non-human primate species.
The researchers found that an average research macaque carried 9.7 million SNVs — more than twice as diverse per individual as humans — and they now plan on using the information to better understand aspects of genome function, such as gene regulation, and to generate new genetic models of disease.
By studying what these nucleotide changes might do to genes implicated in autism spectrum disorders and predicting likely gene disruption (LGD) variants, for example, the macaques may offer new clues to the often heterogeneous genetic condition.
These naturally occurring mutations could provide an opportunity to develop noninvasive models of human disease without the expense of CRISPR engineering of embryos, they added, which would be particularly useful in relation to phenotypes that are not readily reproduced in non-primate knockout models and for evaluating the effect of genetic variation on the efficacy of treatments before human trials.
“This new macaque reference genome and the genetic characterization of research populations will substantially advance biomedical research and studies of primate genome evolution by providing an improved framework for more complete studies of genetic variation and its phenotypic consequence,” the authors concluded.
Explore how to use PacBio whole genome sequencing and the Iso-Seq method to advance your research.
January 28, 2021 | Plant + animal biology