
As we close out PAGBio Day 2022, there is one more key part of genomic natural history to consider – animal genomics.
As we’ve outlined in prior posts, it’s clear that we have entered an era of genomic natural history. Yet a comprehensive playbook where every species has a corresponding, reference-quality genome assembly is not quite ready.
How far have we come, and what is most needed to complete the picture?
A recent PNAS Perspective piece by Paul B Frandsen (@paulbfrandsen) of Brigham Young University, and Washington State University researchers Joanna Kelley (@joannalkelley) and Scott Hotaling (@MtnScience) reviewed the past 25 years of animal genetics, and highlighted disparities in genomic representations, including a systemic overrepresentation of vertebrates and underrepresentation of arthropods.
From the first animal genome sequence published-the 97 million–basepair (bp) (Mb) Caenorhabditis elegans–to the nearly 3,300 unique animal species across 24 phyla that are in GenBank today, our knowledge of how genomes vary and shape Earth’s biodiversity has grown significantly since 1998. And the momentum is increasing, with several sequencing consortia feverishly working towards a collective goal of sequencing all animal genomes, including the Vertebrate Genomes Project, the Bird10K project, the Bat1K project, the i5K project, the Earth BioGenome Project, and the Darwin Tree of Life Project.
Still, there’s a long way to go. As authors point out, only 0.2% of the roughly 1.66 million described species in the animal kingdom currently have their nuclear genome sequenced. However, the pace of sequencing is speeding up, with an average rate of 0.52 species assemblies submitted to the GenBank repository per day increasing eightfold over the past year to 4.07 assemblies per day.
If the most recent rate were maintained, all currently described animals would have a genome assembly available by 3136. To achieve this goal by 2031, an average of 165,614 novel animal genomes would need to be sequenced and assembled each year (∼112 times faster than the rate for the most recent year), the authors note.
Mind the gaps
While celebrating the advances in genomic discovery, we also must acknowledge the disparities that exist so that we can plan for the future. Currently, genome assemblies are available for 685 ray-finned fishes, but none exist for phylum Nematomorpha, an ∼2,000-species clade of parasitic worms whose presence can dramatically alter energy budgets of entire stream ecosystems.
So where are the taxonomic gaps? The publication highlights a few:
- Genome assemblies for 3,278 species representing 24 phyla, 64 classes, and 258 orders. Collectively, 14 groups were underrepresented, 17 were represented as expected,
and 28 were overrepresented. - The phylum Chordata (which includes all vertebrates) had 1,770 assemblies for the group (54% of all assemblies), despite chordates comprising just 3.9% of animal species
- Conversely, arthropods had 1,115 assemblies (34% of the dataset) for a group that comprises 78.5% of animal species.
- Ten phyla had no publicly available genome sequence
The authors warn about the unique biology that is being overlooked, and urge researchers to diversify their sequencing efforts.
“From the perspectives of biomedicine and human evolution, this bias [in favor of vertebrates] is reasonable since humans are vertebrates,” the authors write. “However, from a basic research perspective, particularly as it relates to genomic natural history and an overarching goal to sequence all animal genomes, there is a need to taxonomically diversify sequencing efforts.”
The paper also notes the disparities in the geographic origin of the species sequenced, and the researchers submitting them. Institutions in North America, Europe, and Asia collectively accounted for 95.5% of all assemblies, and nearly 70% of all animal genome assemblies have been submitted by researchers in just three countries: United States (n = 1,275), China (n = 676), and Switzerland (n = 317).
Interestingly, researchers at North American institutions have contributed the most insect and mammal assemblies, European researchers have contributed the most fish assemblies, and Asian researchers have contributed the most bird assemblies.
“Similar to how sampling biases can yield skewed understanding of the natural world in other disciplines, so too could bias toward specific ecoregions, habitats, or other classifications skew genomic insight,” they write.
The authors call for more geographic representation, especially from The Global South, which is home to the bulk of the world’s biodiversity.
“It behooves everyone, including researchers in the Global North, to deepen collaborations with peers in the Global South while also helping to build indigenous capacity for collection, storage, and sequencing of new specimens.”
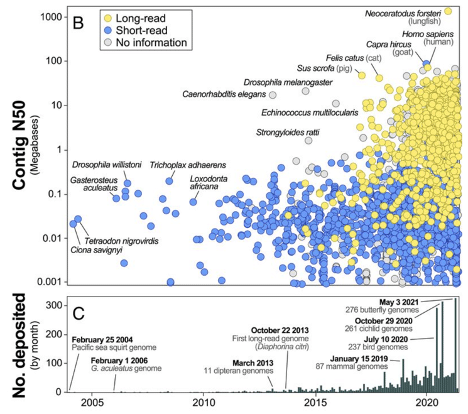
All creatures great and small
From the smallest genome assembly in the dataset, the 32.5 Mb mite Aculops lycopersici, to the largest (nearly 1,000 times bigger) the 32.4 Gb axolotl and the 34.6 Gb Australian lungfish, contiguity also varied dramatically across groups, the authors noted.
Hominid and bird assemblies were the most contiguous with an average contig N50 of 24.2 Mb and 1.4 Mb, respectively. On the other end of the spectrum, jellyfish and related species (phylum Cnidaria) exhibited some of the least contiguous genome assemblies with a mean contig N50 of 0.18 Mb.
“For the purpose of biological discovery, not all genome assemblies are created equal,” they write.
As long-read sequencing technologies have matured, so too has the quality of assemblies being generated. Going forward, the quality of a genome assembly is likely the most important factor dictating its long-term value, the authors state, and they call for future assemblies to echo the guidelines proposed by the Earth BioGenome Project and “exceptional” genetic resources contributed by the Vertebrate Genomes Project and the Darwin Tree of Life.
“Assemblies should reach minimum levels of contiguity (e.g., contig N50 > 1 Mb) and accuracy in order to be considered a reference that will likely not need to be updated for most applications,” they state.
Interested in reading more about long-read sequencing and HiFi reads? Check out our Plant + Animal page to learn more about how they empower insect biology, crop improvements, animal health and breeding and more.
● How Far We’ve Come Part 1: Insect Genomics
● How Far We’ve Come Part 2: Plant Genomics