
Plant genomes are notoriously difficult to assemble. They are large and complex, with varying levels of ploidy. In fact, the genome of one of the most well-known model species, Arabidopsis thaliana, wasn’t even released until 2000.
Luckily, advances in long-read sequencing technology and new computational tools have made sequencing and assembly of virtually any species possible. New plant genomes are rapidly making their way into genetic repositories, with more than 800 to date. Overall, 74% of land plant genome assemblies have been produced in the past three years.
But, there is still much further to go. There are disparities in taxonomic diversity and geographic representation that HiFi sequencing technology can help overcome.
A review in Nature Plants by Michigan plant scientists Rose Marks (@RoseAMarks1) and Robert VanBuren (@bob_vanburen), along with co-authors Scott Hotaling (@MtnScience) of Washington State University and Paul B Frandsen (@paulbfrandsen) of Brigham Young University, identified numerous taxonomic gaps and disconnects between the native range of focal species and the national affiliation of the researchers studying them.
The first land plants to have their genomes sequenced and assembled were model or economically important crop species with small diploid genomes. This is not surprising, as until recently, technological limitations made it difficult to assemble high-quality polyploid genomes.
That’s where HiFi sequencing comes in.
“With the improvements offered by long-read sequencing, it is becoming more feasible to sequence and assemble large-polyploid plant genomes,” the authors note.
As evidenced by the sequencing of the giant California redwood (Sequoia sempervirens) and the assembly of its ginormous 27 Gb genome, what would have been considered a herculean effort not that many years ago was accomplished in only a few weeks by a tiny team of scientists in their spare time.
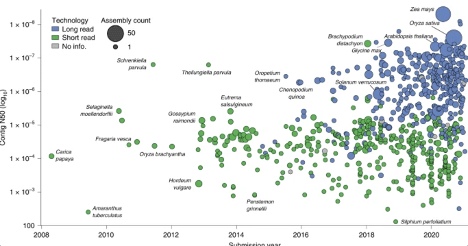
Assembly contiguity by submission date for 798 land plant species with publicly available genome assemblies. Points are colored by the type of sequencing technology used and scaled by the number of assemblies available for that species.
The urgency to assemble native and wild species has never been greater, with climate change and conservation becoming a top priority. As PacBio CSO Jonas Korlach noted in the blog, more biodiversity means more resilient ecosystems, and every species–ours included–will benefit from such research.
Marks, et al., also noted the need to learn what we can from wild species before they disappear; plant extinction has already increased 60% over the past 100 years, and this is projected to continue even under the most optimistic scenarios.
“We urge researchers to take advantage of new genomic technologies that provide an opportunity to explore, catalog and mine the immense diversity of information contained within wild species before they are lost.”
Minding the gaps for future discovery
While the understanding of plant genomics has advanced greatly in recent years, there are many taxonomic gaps that offer the opportunity for exploration. Here are a few significant taxonomic gaps to note:
Of the 137 land plant orders that have been described, over half (76) lack a representative genome assembly.
- Genome assemblies are available for 135 domesticated, 127 cultivated, 120 natural commodity and 12 feral species. The remaining 404 genome assemblies are from wild species; of these, 77 are wild relatives of crops.
- Six orders of land plants are statistically over-represented in genome assembly databases based on species richness, including the agriculturally and economically important clades of Brassicales, Cucurbitales, Fagales, Malvales, Rosales and Solanales.
- Four orders of land plants had significantly fewer genome assemblies than expected based on species richness: Asparagales, Asterales, Gentianales and Polypodiales.
- Bryophytes are poorly represented, with assemblies for only eight mosses, three liverworts and three hornworts.
“While the number of human-linked species with genome assemblies is largely equivalent to wild species, this equivalence reflects an extreme bias,” the authors note. “There are far more wild (~350,000) than domesticated species (~1,200–2,000), suggesting that wild plants represent an untapped reservoir of genomic information.”
And while many plant genome assemblies are for species that are native or economically important in Africa and South America, chances are they were sequenced and assembled by researchers elsewhere.
In fact, the researchers found significant geographic gaps, with ~77% of genome assemblies attributed to a handful of affluent nations primarily from the Global North: China (235 assemblies), the United States (212) and Europe (168). Research teams in Oceania, South America and Africa published just 40, 9, and 1 assembly, accordingly.
They estimate that 56% of all domesticated crops have had their genome sequenced outside of their continent of origin and only 13% of these included in-continent collaborators.
“This represents a substantial global imbalance in genomics,” the authors state.
Less input from local stakeholders means increased likelihood that genome assemblies may not actually represent the germplasms grown there, or conservation priorities in those regions.
“We encourage all plant scientists to strive to support local stakeholders, to incorporate indigenous knowledge into their work and to invest in building systems and expertise for working with genomic resources in the location where they occur naturally,” the authors write.
How to fill the gaps as we move forward
While many of these gaps can be attributed to historic disparities in scientific and economic wealth, technological advances could help democratize access and participation, the authors suggest.
“Falling sequencing costs, widening availability of analytical tools and an increasingly connected scientific community provide key opportunities to improve existing assemblies, fill sampling gaps and empower a more global plant genomics community,” they write.
In terms of disparities in data quality, the authors also call on plant genome scientists to embrace long-read sequencing technologies and leverage them whenever possible to generate new assemblies.
“This is already occurring but, given the massive disparity in quality between assemblies generated with short-read versus long-read data, the need for continued adoption cannot be overstated,” they write.
Interested in reading more about long-read sequencing and HiFi reads? Check out our Plant and Animal page to learn more about how they empower insect biology, crop improvements, animal health and breeding and more.