It’s time to revisit the way scientists are using 16S rRNA gene sequencing to study microorganisms, according to a team of Jackson Laboratory researchers.
Popular targets for taxonomy and phylogeny studies because of their highly conserved nature, amplified sequences of the 16S ribosomal RNA genes can be compared with reference databases to determine the identity of the microorganisms that comprise a metagenomic sample. Sequences with a > 95% match are generally considered to represent the same genus, for example, while > 97% matches are considered the same species.
However, these matches are often made by sequencing only part of the nine-region, ~1500 bp 16S gene, either single regions like V4 or V6, or variable regions like V1–V3 or V3–V5, as done in the Human Microbiome Project. In a paper published recently in Nature Communications, Jethro S. Johnson, George M. Weinstock and colleagues point out that it is time to revisit this compromise that arose only because of past technological limitations. Given recent advances in long-read sequencing accuracy, the entire 16S gene should now be interrogated, the authors suggest.
Circular consensus sequencing (CCS, the method used in PacBio HiFi Sequencing), in particular, combined with sophisticated denoising algorithms, means it is now possible to sequence the entire gene with sufficient accuracy to discriminate among millions of sequence reads that differ by as little as one nucleotide, they write.
“Together, these technological and methodological advances mean that for the first time, it is becoming possible to exploit the full discriminatory potential of 16S in a high-throughput manner,” the authors write.
Using an in-silico dataset of 16S sequences taken from the Human Microbiome Project database, the researchers demonstrated that commonly targeted sub-regions were unable to recapitulate the taxonomic information present in the full 16S gene.
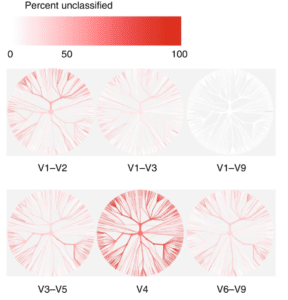
“The V4 region performed worst, with 56% of in-silico amplicons failing to confidently match their sequence of origin at this taxonomic level,” they wrote.
“Our simple in-silico experiment demonstrates that it is not valid to assume that ever finer clustering of these sub-regions will result in the improved taxonomic resolution necessary to reflect species.”
They also found that different sub-regions showed bias in the bacterial taxa they were able to identify at the species level. For example, while V1-V3 gave good results for Escherichia and Shigella, good results for Klebsiella required the V3-V5 region, whereas Clostridium and Staphylococcus required V6-V9 sequencing. Since all of these strains may be present in the human gut, the only way to ensure good taxonomic identification of all species is to sequence the full gene from V1-V9.
However, the team points out that it may be possible to obtain even better taxonomic resolution, down to the strain level. Bacteria have between 1-15 copies of the 16S genes. While the number of copies is consistent within a species, the intragenomic variation among the copies is strain specific. The Jackson Lab team believes this intragenomic variation presents an opportunity.
For example, sufficient nucleotide variation exists to distinguish E. coli strain K-12 MG1655 from the infection-causing O157 Sakai strain. The team provides proof of concept evidence for this approach with full-length PacBio 16S sequencing data from 381 isolates selected from the Human Microbiome Project sample bank. They show that the vast majority of these bacteria can be uniquely assigned to a specific strain using the intragenomic 16S variation revealed by PacBio 16S HiFi data.
“Thus, we argue that, when appropriately accounted for, multiple polymorphic 16S copies are not an inconvenience to be overlooked, rather they will enable the 16S gene to be used in strain-level microbiome analysis,” they add.
“Analysis of microbial communities at these taxonomic levels promises to provide a very different perspective to the one afforded by genus-level abundance estimates.”
Learn more about the methods and workflow for PacBio full-length 16S sequencing.
December 11, 2019 | Microbial sequencing methods