
It’s been a year since we took a little field trip to Stanford to collect samples from the giant California redwood (Sequoia sempervirens) with the goal of assembling its ginormous 27 Gb genome.
What would have been considered a herculean effort not that many years ago was accomplished in only a few weeks by a handful of personnel —Emily Hatas (@EmilyHatas), Greg Young (@PacbioGreg), Michelle Vierra (@the_mvierra), and Greg Concepcion (@phototrophic) — in their spare time.
As detailed in this blog post, the crew put together an assembly with 22-fold coverage in just 17 days — 4 days of sample prep, 7 days of sequencing, and 6 days for assembly. With a little more time on their hands, and enough HiFi library to go around, the team embarked on more sequencing to create an even better assembly, with 33-fold coverage, a contig N50 of 3.8 Mb.
Wanting to take things even further, Iso-Seq analysis expert Elizabeth Tseng (@magdoll) delved deep into sequences of transcripts from the redwood’s needles.
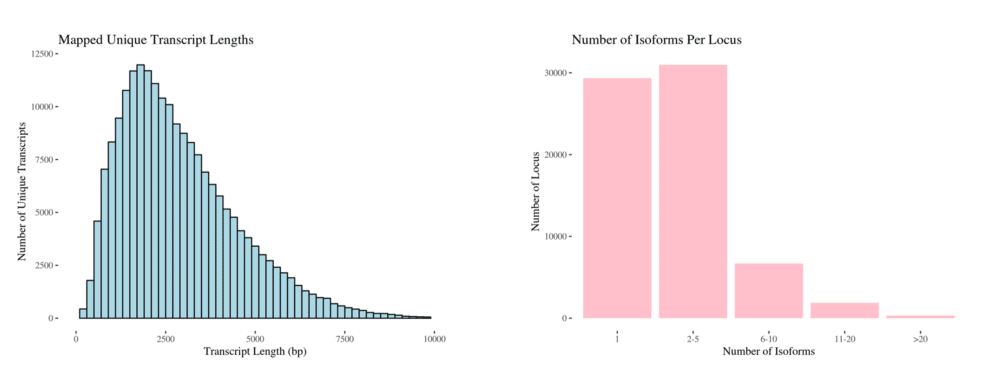
The results from two Sequel II SMRT Cells (a total of 5.3 million full-length reads) were mapped to the hifiasm v12 assembly of the PacBio redwood genome, yielding 336,853 high-quality Iso-Seq transcripts, with 69,198 mapped loci and 205,792 unique, full-length transcripts.
The mapped transcripts ranged from 50 bp to 14.2 kb with a mean length of 2.9 kb. While most of the loci had 1–5 isoforms, there were many that displayed complex alternative splicing patterns, highlighting the power of full-length transcript sequencing.
“I found several aspects of the Iso-Seq data exciting,” Tseng noted in a Medium post about the work. “One was the ability to see alternative splicing. Another was the ability to predict ORFs directly from the sequences.”
The exercise was also a good test of the IsoPhase isoform phasing method that Tseng initially developed for maize, a diploid genome. Would it work for a hexaploid genome?
Yes.
By combining phased genome information with phased transcriptome data, Tseng was able to identify five distinct alleles, as well as genes that were likely to be homologous.
Lastly, Tseng used the Iso-Seq data — and another tool, Cogent — to assess the quality of the redwood genome assembly.
“The high mappability of the Iso-Seq data to the PacBio genome has shown that the genome assembly is quite complete in terms of coding regions,” she said. “Missing genes or difficult-to-assemble gene regions can be assessed using Iso-Seq transcripts.”
Want To See More Redwood Iso-Seq Analysis? Dig In! We’ve released the Iso-Seq dataset, including the transcript sequences, GFF files, BLASTN hits, IsoPhase and Cogent results. We welcome the community to use this dataset for research, tool development, and give us feedback.
Data for the redwood genome has also been made publicly available along with the updated genome assembly and can be found here.
—
Interested in finding out more about HiFi data for sequencing your organism of interest? Get in touch with a PacBio scientist to scope out your project.
March 4, 2021 | Plant + animal biology