How do bacteria manipulate plant biology to cause blight and rot? Why are some pathogen strains more virulent than others? How can we engineer resistant staple food crops? These are pressing questions facing researchers looking to sustain and increase crop production against the backdrop of a changing environment.
For one major clade of pathogens, Xanthomonas spp, the answers lay locked within TAL effector genes (TALEs), but assembling these highly variable, repetitive regions was a long-standing obstacle. The key to finally unraveling the tangled assemblies was PacBio long-read sequencing.
Code-breaker

Plant pathologist Adam J Bogdanove (@AdamBogdanove) and colleagues at Cornell University have been using SMRT Sequencing to elucidate the structure and function of TALEs and generate new insights into the mechanisms of Xanthomonas virulence.
“Repeats render TAL effector genes nearly impossible to assemble using next-generation short reads… long-read, single molecule real-time (SMRT) sequencing solves this problem,” Bogdanove wrote in one study.
Bogdanove was one of two researchers to break the TALE-DNA code in 2010 by searching for patterns in protein sequence alignments and the promoter sequences of genes upregulated by TALEs.

What are TALEs?
TALEs are proteins secreted by Xanthomonas bacteria when they infect various plant species (including pepper, rice, citrus, cotton, tomato, and soybeans), causing localized leaf spot and leaf streak, or systemic black rot and leaf blight disease.
How do they work?
TALEs bind promoter sequences in the host plant and activate the expression of plant genes that aid bacterial infection. They recognize plant DNA sequences through a central repeat domain consisting of a variable number of tandem ~34 amino acid repeats that vary at only 2 positions. These two critical amino acids correspond to a DNA base in the target promoter sequence. Bogdanove figured out that HD binds to C, NI to A, NH to G, etc.
The enabling technology
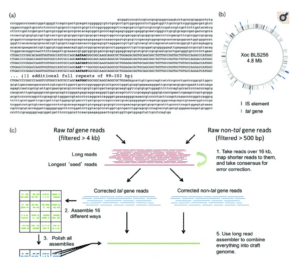
How did Bogdanove come to rely on SMRT Sequencing for TALE research? In a 2015 paper in Microbial Genomics, he described comparing PacBio assemblies to existing Sanger-based reference genomes of X. oryzae pathovars. The exercise revealed errors and omissions in the Sanger sequences, and the team concluded that PacBio sequencing was the best tool for generating de novo, whole-genome assemblies for Xanthomonas that accurately capture TALE genes.
Accurate assembly of TALE genes can be ensured by pre-assembling reads that contain TALE gene sequences. Together with co-author Nicholas J. Booher and others at Cornell, Bogdanove created a workflow for TALE gene assembly and prediction of their target sequences, the “pbx toolkit,” available on Github. Since then, the Bogdanove lab, along with other researchers worldwide, have used SMRT Sequencing to elucidate the molecular mechanisms of TALE function and the dance of susceptibility and resistance between pathogen and host.
Tracking virulence

Bogdanove and colleagues have used PacBio sequencing on Xanthomonas strains that infect important food crops worldwide. He writes, “Identification of the complete sets of TALEs in different isolates can be an important first step toward development and targeted regional deployment of resistant soybean varieties, and comparison of whole genome structure across strains can yield insight into the overall genetic diversity of the pathogen.”
A 2019 Genome Biology and Evolution paper describes the complete assembly, including all TALE genes, of Xanthomonas axonopodis pv. glycines isolates collected from infected soybean plants. Surprisingly, they found that the TALEs of the three strains they sequenced were highly similar, despite having been collected over a span of 30 years and on two different continents. Bogdonove concludes that if there is “little to no genetic variation at their targets across commonly grown soybean varieties, such that there is no selective pressure on the tal genes to adapt,” these genes may be good targets for the development of resistance.
In another paper published in Frontiers in Microbiology, Bogdanove and colleagues in Iran used SMRT Sequencing to better understand the role of TALEs in the virulence of bacterial leaf streak in wheat caused by Xanthomonas translucens pv. undulosa (Xtu).
They sequenced the genome of the highly virulent Iranian strain ICMP11055, generating a closed 4.5 Mb genome. They then compared it to the XT4699 strain from the United States, finding two major re-arrangements, nine genomic regions unique to ICMP11055, and one region unique to XT4699, as well as differences in TAL effector genes. Mutagenesis and complementation experiments indicated that at least a subset of the TALEs contribute to the virulence of these strains in wheat.
“Our results lay the foundation for identification of important host genes activated by Xtu TALEs as targets for the development of disease resistant varieties,” the authors wrote.
Tracking the SWEET tooth of TALEs
Finding the target genes of TALEs in plant hosts is critical to understanding the co-evolution of bacterial virulence and plant resistance. In a Nature Communications paper, Bogdanove and his team used PacBio sequencing to link Xanthomonas citri subsp. malvacearum (Xcm) TALEs to SWEET (‘sugars will eventually be exported transporter’) target genes in cotton. By correlating cotton transcriptome profiling with Xcm TALE DNA binding site prediction, they postulated a connection between TAL effector Avrb6 and the induction of sucrose transporter GhSWEET10.
In follow-up experiments, the authors found that “activation of GhSWEET10 by designer TAL effectors (dTALEs) restores virulence of Xcm avrb6 deletion strains, whereas silencing of GhSWEET10 compromises cotton susceptibility to infections.” “These findings advance our understanding of the disease and resistance in cotton and may facilitate the development of cotton with improved resistance to BBC.”
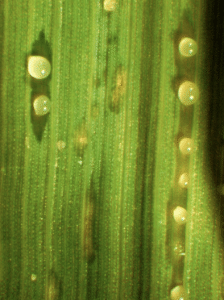
Another study of SWEET genes focused on host resistance to the rice pathogen Xanthomonas oryzae pv. oryzae (Xoo). SWEET activation by TALEs leads to sucrose export into the xylem vessels, facilitating Xoo proliferation in rice. Researchers have identified and cultivated 42 bacterial blight resistance genes in rice, called Xa genes, which can prevent binding and activation by the cognate TALEs via two distinct mechanisms, reducing susceptibility to Xoo. All but one of the resistance genes are SWEET alleles that lack binding sites for TALEs. The final resistance gene is a mutation in a transcription factor gene that prevents TALE binding to the plant transcriptional machinery.
However, bacterial strains that can defeat these resistance mechanisms have arisen in India and Thailand. So, together with collaborators in those countries, Bogdanove sequenced the genome of one such strain from each country. While examination of the encoded TALEs revealed how the Xoo strains escaped the protective mutation of a key transcription factor gene, the mechanism for escaping protective mutations in SWEET alleles remained unclear but the data suggested an experimental path for resolving the mystery.
“The findings open a door to mechanistic understanding of the role SWEET genes play in susceptibility and illustrate the importance of complete genome sequence-based monitoring of Xoo populations in developing varieties with effective disease resistance,” the authors wrote.
Re-evaluating references
SMRT Sequencing also helped correct a long-held belief regarding Brassicaceae-infecting Xanthomonas campestris (Xc). TALE-encoding genes were thought to be absent from Xc genomes based on four reference genomic sequences. But as reported in New Phytologist, Bogdanove and colleagues from the Université de Toulouse discovered TAL genes in 26 of 49 Xc strains isolated worldwide.
Using a combination of SMRT and TALE amplicon sequencing, they created a “TALome,” a near-complete description of the TALEs found in Xc. The new resource will “open novel perspectives for elucidating TALE-mediated susceptibility of Brassicaceae to black rot disease,” the authors wrote.
The complexity of bacterial genomes
While bacteria have a reputation for having small and tractable genomes, in truth there are many clades where the presence of numerous genes from highly repetitive gene families is common. Adding to the assembly complexity, these genes are often flanked by similarly repetitive mobile elements. PacBio sequencing offers a simple, affordable solution to closing even the most challenging bacterial genomes, enabling new insights into key biological processes.
Learn more about microbial whole genome sequencing, connect with a service provider, or talk to a PacBio scientist.
August 19, 2020 | Plant + animal biology