
May is ALS Awareness Month, and we’re hoping to help raise awareness by shining a spotlight on two deserving publications from scientists at the University of Washington and at the Mayo Clinic. In both studies, researchers used PacBio technology to sequence targeted genomic regions and to discover and characterize complex pathogenic variants associated with ALS.
ALS, short for amyotrophic lateral sclerosis and commonly known as Lou Gehrig’s disease, is a progressive disease of the nervous system that causes loss of muscle control. The mean survival time for patients with ALS is just three to five years from diagnosis, and there is no cure for the disease.
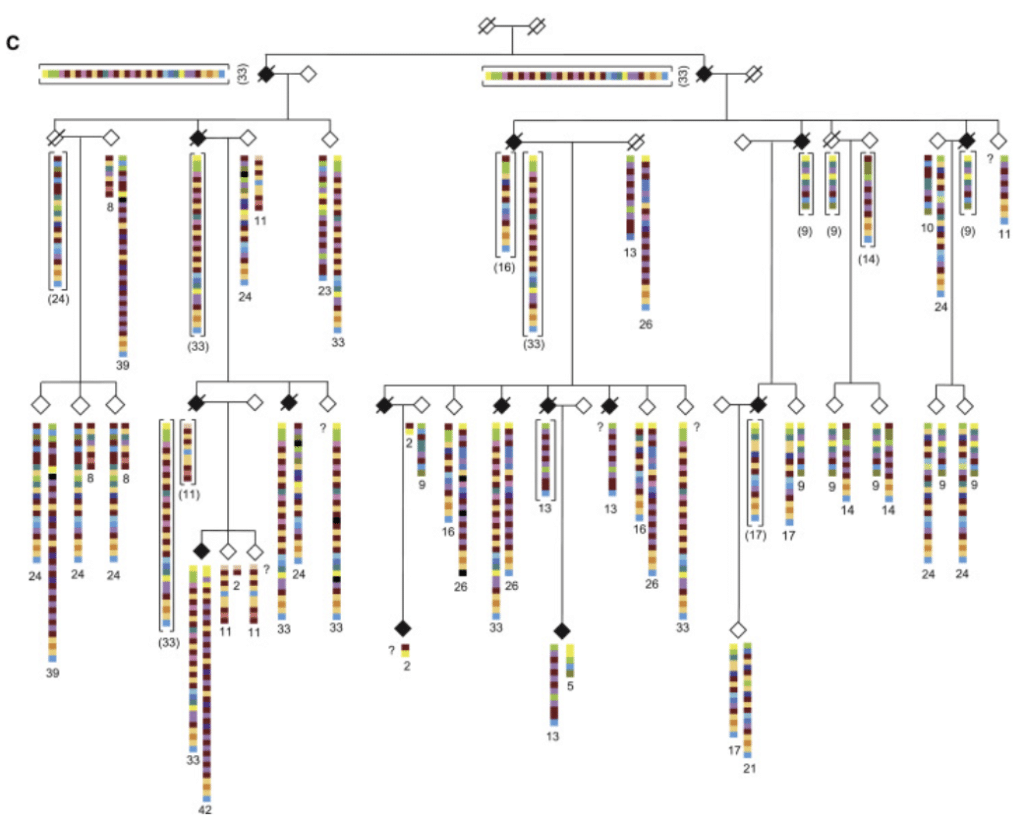
At the University of Washington, scientists studied members of a large multigenerational family in which several people developed spontaneous ALS. By using barcoded amplicons and a multiplex strategy, the team analyzed the target WDR7 gene in nearly 300 samples and identified a variable number tandem repeat (VNTR) that is expanded in individuals with ALS.
Their paper, published in the American Journal of Human Genetics, details single-base resolution for a repeat that would have been virtually impossible to spot — let alone resolve with such granularity — using any other sequencing technology. “Our detailed interrogation of this VNTR demonstrates the value of high-depth, long-read sequencing of human-specific repetitive regions that expand in the genome,” the team reports.
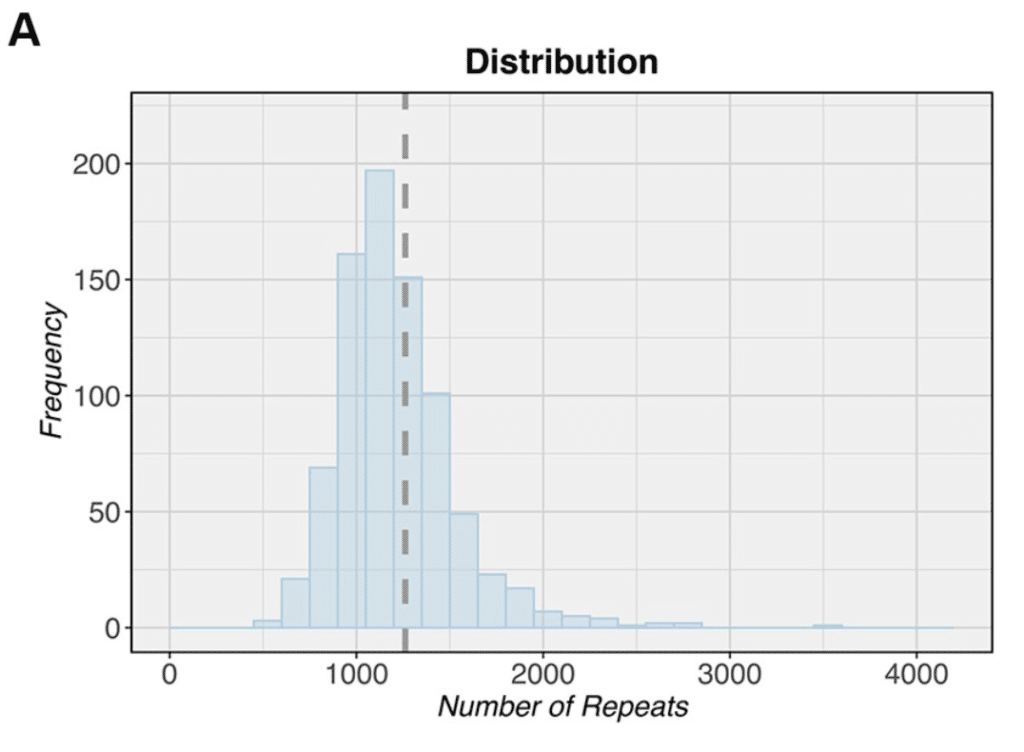
Meanwhile, at the Mayo Clinic, scientists used PacBio’s CRISPR/Cas9-based No-Amp method to target, capture, and sequence a repeat expansion region associated with neurological diseases. The targeted element is the most common genetic cause of ALS, but the biological mechanism of how it causes disease has not been well understood. The team used PacBio sequencing to characterize the complete region, finding that individuals with fewer repeat copies had longer survival time than those with larger expansions. Results were recently published in the journal Brain.
“Our findings demonstrate that No-Amp sequencing is a powerful tool that enables the discovery of relevant clinicopathological associations, highlighting the important role played by the cerebellar size of the expanded repeat in C9orf72-linked diseases,” the scientists write.
These discoveries could open up new paths to identify people at risk of developing ALS. Congratulations to both of these teams, as well as other PacBio users who are making a difference for families affected by ALS!
Interested in learning more about our technology and ALS research?
- Read our newest groundbreaking case study: ‘How SMRT Sequencing Helped Researchers at University of Washington Uncover a Tandem Repeat Linked to ALS’
- Watch Marka van Blitterswijk from the Mayo Clinic present, ‘Applying Targeted Long-read Sequencing to Assess an Expanded Repeat in C9orf72’
- See Meredith Course from the University of Washington present, ‘The Evolution and Function of a Large Tandem Repeat Associated with ALS’.
Visit our Neuroscience Research page to learn how PacBio sequencing provides a comprehensive understanding of the genetic basis of neurological disease.