Unlocking the secrets of complex communities and microbiomes represents one of the most exciting frontiers in microbiology, with the potential to revolutionize our understanding of human health, food production, and environmental preservation. To lay the groundwork for such breakthroughs, fully capturing and profiling all the genetic information present in a microbiome or complex community is critical; and shotgun metagenomic methods are essential for obtaining such insights.
In pursuit of the best shotgun metagenomic data, PacBio HiFi sequencing continues to set the standard for quality and completeness, thanks to exciting innovations in data analysis tools. These new pipelines, paired with HiFi data’s potent combination of high-accuracy and long read-length, yield more precise profiling information than any other technology on the market. Moreover, HiFi data returns more circular, single-contig metagenome assembled genomes (MAGs) than ever before, giving you the power to uncover hidden microbial ecosystem functions and confidently capture species richness and diversity.
- HiFi shotgun metagenomic sequencing generates datasets made of individual reads spanning up to 8 contiguous genes, allowing you to precisely profile the function and taxonomy of a community, all at once.
- Taxonomic and functional profiling of HiFi metagenomes can now be performed at nearly 1/3rd the cost† without compromising accuracy, thanks to insights from downsampling experiments.
- Assembly-focused HiFi metagenomic studies can return tens to hundreds of high-quality (HQ) MAGs with up to 33% of those MAGs being single contig, allowing you to predict genes and make taxonomic annotations with authority.
Obtain HiFi metagenomic profiling data of stunning quality, at a lower cost
Profile more affordably using only 0.5 Gb of HiFi data to obtain the same state-of-the-art species and diversity information that can be found with 88 Gb, or presumably more.
The proof is in the research: In order to help microbiologists obtain the most cost-effective insights possible from HiFi data, we performed a series of downsampling studies. Each analysis was conducted to find the minimum depth necessary to achieve very accurate community profiles as well as determine HQ and single-contig MAG recovery rates.
To begin, we sequenced the ZymoBIOMICS fecal reference with TruMatrix technology, which is a pooled highly complex human gut microbiome sample that is extremely useful for benchmarking and reference standards. We sequenced this sample using 4 SMRT Cell 8Ms and then took the dataset and downsampled it incrementally to a coverage equivalent with a 96 multiplexed sequencing run on a single SMRT Cell 8M(96-plex). The study captured a total range of 88 to 0.3 gigabases (Gb) of data. Each data level was then run through PacBio’s taxonomic profiling workflow.
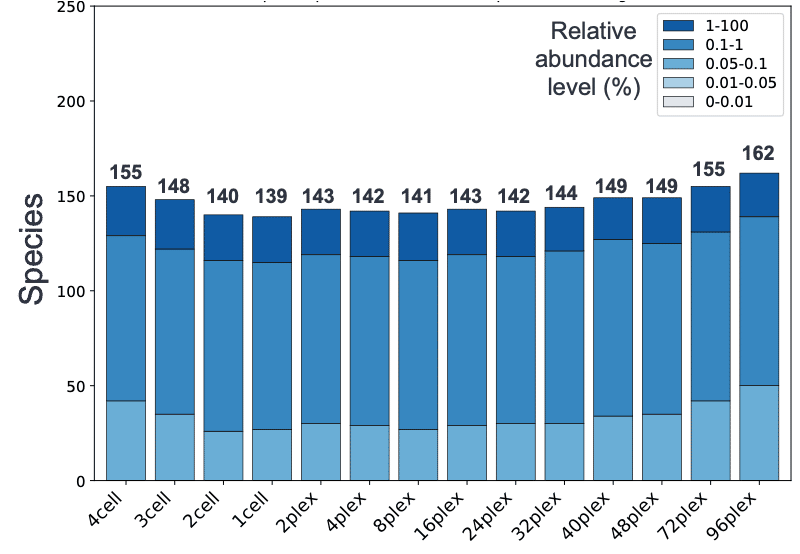
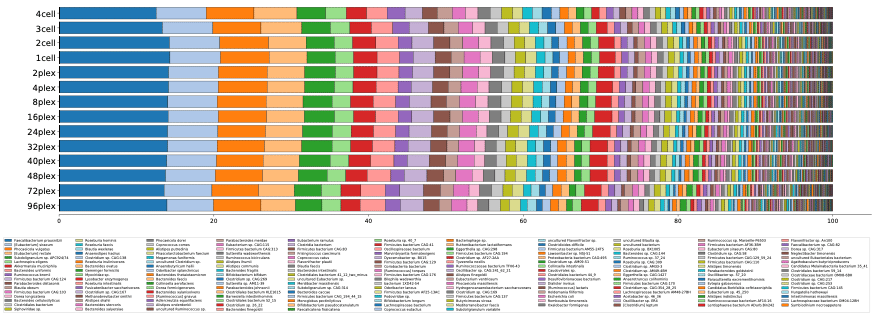
The results from this workflow showed that the information obtained from 4 SMRT Cell 8Ms down to 48-plex is largely consistent, with not only similar numbers of species being recovered (fig. 1), but the relative abundance profiles being nearly identical as well (fig. 2). This indicates that the same level of taxonomic profiling information can be obtained with 0.5 Gb at the 48-plex level as 88 Gb, offering microbiome researchers an even more affordable entry point into the world of HiFi metagenomics.
Listen to Dan Portik PhD (@DPortik) Senior Scientist, Bioinformatics at PacBio discuss these results in our recent metagenomics webinar, on demand now.
Capture species in exquisite detail with more HQ-MAGs than ever
Achieve optimal cost-benefits starting at one sample per SMRT Cell 8M for assembly-focused metagenomic HiFi sequencing. Projects seeking greater insight can increase sequencing depth with multiple SMRT Cell 8Ms to capture rare microbes in vivid detail.
How we know: The second half of our downsampling study was dedicated to establishing guidelines for metagenome assembled genome (MAG) focused studies. To assess the effects of sequencing depth on MAG assembly, we took the ZymoBIOMICS TruMatrix 4 SMRT Cell 8M gut microbiome dataset and incrementally downsampled it to an 8-plex depth to look at the effects on assembly and recovery at each level. We know that assembly is much more sensitive to sequencing depth and that there is a very predictable relationship between sequencing depth and the outcome. However, in our pipeline, depth impacts HQ-MAG recovery and single-contig MAG recovery differently.
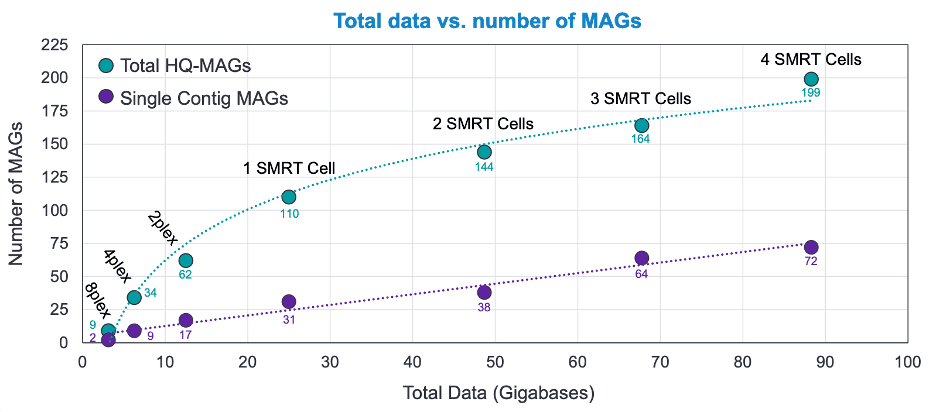
The results from our workflow show that from 8-plex on there’s a log relationship between total MAG recovery and depth (green line in fig. 3), with rapid gains up to a single SMRT Cell 8M. From here, the efficiency of recovery is somewhat reduced as sequencing depth increases but HQ-MAGs are still returned in large numbers with the deepest sequencing yielding what are almost certainly rarer species. For the recovery of single-contig MAGs (fig. 3, purple line), there is a distinct linear recovery-to-depth relationship with 4 SMRT Cell 8Ms yielding 199 total HQ-MAGs, 72 of which are single contig. Even at a depth of 8-plex, 9 HQ-MAGs were recovered and 2 were single contig. At 4-plex, 34 HQ-MAGs were generated with 9 being single-contig. These results clearly indicate that HiFi shotgun metagenomic sequencing can deliver numerous and strikingly detailed genome assemblies that would be difficult to obtain with alternative sequencing technologies.
Listen to Dan Portik PhD (@DPortik) Senior Scientist, Bioinformatics at PacBio discuss these results in our recent Metagenomics Webinar, on demand now.
Peer into the microbial world with astounding clarity
HiFi metagenomics gives microbiome researchers the power to precisely profile the function and taxonomy of a community with greater cost-effectiveness than ever before. At the same time, the HiFi reads’ incredible length and accuracy allow investigators to leverage impressive HQ-MAG recovery to predict genes and make taxonomic annotations with greater confidence. Together these advantages make HiFi sequencing the premier sequencing solution for ambitious projects aimed at groundbreaking discoveries.
Hungry for more?
Check out the recap from our recent metagenomics webinar
Watch our HiFi metagenomics talk, available on-demand now
Check out our HiFi solutions for metagenomics
Access HiFi metagenomics datasets
Try out the PacBio HiFi metagenomics pipeline