We recently brought together Jeremy Wilkinson, Ph.D. (@jewilki), Microbiome and Metagenomics Market Lead at PacBio, Xiaowen Feng, Ph.D. (@0xfxfxf), Postdoctoral Fellow at Dana-Farber Cancer Institute and Harvard Medical School, and Daniel Portik, Ph.D. (@DPortik), Senior Bioinformatics Scientist at PacBio to discuss recent advances in the PacBio HiFi metagenomic data processing pipeline.
During the webinar, it was shown that, when combined with PacBio HiFi sequencing, this new suite of analysis tools delivers data of unparalleled quality and breadth, enabling microbiologists to achieve a never-before-seen level of taxonomic detail and functional insight into microbial communities.
Key Takeaways:
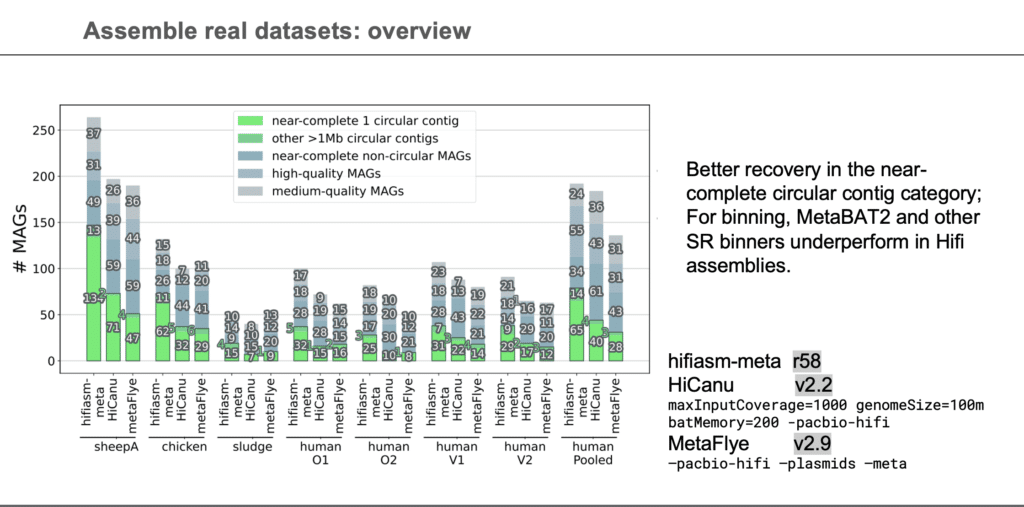
- The newly developed hifiasm-meta software tool enables the PacBio HiFi metagenomics pipeline to correctly identify more circular single-contig MAGs and more total MAGs than any other process available.
- PacBio HiFi sequencing reads enable metagenomic studies to achieve taxonomic resolution to the level of species, and even strain, in-tandem with functional insights.
- HiFi metagenomic down-sampling experiments show near-equal data breadth from one sample run on 4 SMRT cells to 48 multiplexed samples in one, enabling you to maximize cost-efficiency while getting state-of-the-art insight from your samples.
“HiFi reads allow us to generate a nearly complete picture of the metagenome, not just a fragmented assembly” -Bickhart et al. Nature Biotechnology, 2022
Hifiasm-meta enhances the power of HiFi metagenomics
The talk began with an exciting introduction about recent advances in shotgun metagenomics. Xiaowen Feng noted how de novo assembly of metagenome samples is a common approach to the study of microbial communities. At the same time, she pointed out how the clear limitations of short-read assemblies and their binning during analysis spurred the development of metaFlye, the only published assembler specialized for long-read metagenome assembly. However, in order to leverage the full power of long, accurate, HiFi reads, Xiaowen Feng and colleagues developed hifiasm-meta, extending earlier work to metagenomic samples. Evaluated against seven empirical datasets, hifiasm-meta reconstructed tens to hundreds of complete circular bacterial genomes per dataset with some >1Mb, consistently outperforming other metagenome assemblers.
Read the article
Get sample data
Try out hifiasm-meta
HiFi metagenomics can capture both taxonomic and functional insights into microbiomes with great precision, simultaneously
Daniel Portik demonstrated the power of the newly optimized PacBio bioinformatics pipeline using 4 pooled human gut microbiome samples from the BioCollective. Taxonomic and functional profiling were performed, as well as metagenome assembly, using analyses tailored specifically to HiFi reads. With taxonomic profiling settings intended to optimize high precision and recall, 199 species were detected across the 4 samples. With less stringent profiling settings, as many as 690 total species were detected. Hifiasm-meta was used to perform metagenome assembly in conjunction with the PacBio binning pipeline to identify and characterize high-quality (HQ) metagenome assembled genomes (MAGs). The hifiasm-meta enabled workflow identified 299 HQ MAGs (>70% complete, <10% contamination, <20 contigs) total across 4 samples. Of these, 141 MAGs composed of a single, circular contig. Lastly, data were downsampled to simulate several multiplexing schemes and investigated the impact on these analyses. Species detection and functional profiling results were robust from 4 SMRT Cells to 48 multiplexed samples in one.
Read the article
Get sample data
Try the PacBio HiFi metagenomics pipeline
Q&A session highlights
Question: Is there a ‘decent’ program for MAG binning using HiFi reads?
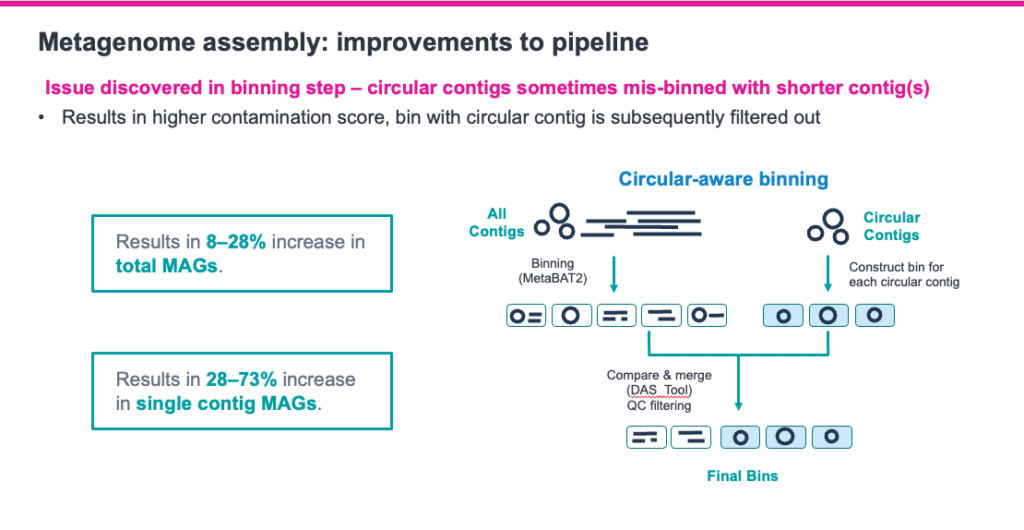
Answer: MetaBAT2 seems to be the best binning tool for HiFi reads thus far, based on testing, but we suggest trying all binners to see what gives the best results for your data. There is still some work that needs to be done with binning of HiFi assemblies.
Question: For read-based profiling, have we done any comparison with that from short reads?
Answer: There are two recent papers that have done these comparisons, Gehrig et al 2022 and Portik et al 2022. With regards to the number of species detected they are comparable after filtering with similar results and consistent species. PacBio reads perform better than short reads in precision and recall and this is because the kmer approaches are not as accurate and even after filtering we can’t achieve the same level of precision and recall that we can get with HiFi reads directly from the pipeline described. More PacBio reads are assigned to functions with typically ~80-90% of reads being annotated, compared to ~1/3 of short reads being assigned.
Question: Most of the results focused on prokaryotic communities. For metagenomic communities which contain eukaryotes, how do you predict HiFi reads will perform in these cases?
Answer: Priest et al 2021 is a good case study demonstrating this type of environment.
Question: For metagenomic assembly, are closed genomes from more abundant organisms or do you also recover single contig MAGs for low abundance organisms?
Answer: Both. It’s easier to close the genomes of the more abundant organisms, but also possible to close those that are lower abundance at as low as 5-10 fold genome coverage and both ends are well represented in multiple datasets analyzed.
Get better results with Pacbio HiFi shotgun metagenomics
HiFi metagenomics promises microbiologists investigating microbial communities a whole new level of data quality, comprehensiveness, and insight. Thanks to newly optimized data processing workflows empowered by hifiasm-meta, researchers can correctly identify more circular single-contig MAGs and more total MAGs than any other process available. PacBio HiFi sequencing reads can achieve taxonomic resolution to the level of species and even strain in-tandem with functional insights while maximizing cost efficiency.
If you missed the webinar or want to revisit any of the key takeaways, watch the full webinar here. Interested in learning more? Visit our metagenomics page.