Since the inception of next generation sequencing (NGS) more than a decade ago, short-read sequencing accuracy has seen only marginal improvement. Having achieved a level of precision thought to be “good enough” for most applications, much of NGS development has been focused on optimizing for cost and throughput. As a result, the underlying raw read accuracy of short-read sequencing (1 in 1,000 bases or Q30) along with its ability to enable important discoveries has all but plateaued… until now.
The introduction of PacBio sequencing by binding (SBB) technology on the Onso short-read sequencing system allows you to generate extraordinarily accurate reads with an error rate of only 1 in 10,000 bases or less (Q40+)!
SBB represents the fundamental breakthrough in short-read sequencing that has long been needed to enable researchers to continue to push the boundaries of what is possible with genomics. But what does this mean for sequencing in general, and how did we get here? Join us as we dive into what SBB sequencing is, how it works, and what its disruptive capabilities will mean for the future of research with short-read sequencing.
The difference is in the chemistry
In contrast to the major strides seen in long-read sequencing technology in recent years, legacy short-read sequencing has seen few changes made to its core sequencing by synthesis (SBS) chemistry. As a result, many research initiatives and methodologies are limited by the accuracy constraints created by this older technology’s underlying chemistry. However, the revolutionary new chemistry that sits at the heart of PacBio SBB technology has the power to completely upend these limitations.
Unlike traditional SBS technology, SBB sequencing uses optimized conditions for each phase of the sequencing cycle, nearly eliminating raw read errors in their entirety. Though seemingly subtle, this critical difference in chemistry constitutes the breakthrough in short-read sequencing accuracy that we have all been waiting for.
What is SBB sequencing?
PacBio SBB sequencing is a Q40+ short-read sequencing technology that works by measuring the light signals from fluorescently labeled nucleotides when they are bound –but not incorporated– by a polymerase on a DNA strand. A computer then matches these light signals with a corresponding nucleotide base call. Unlike other short-read technologies, SBB separates the binding and subsequent extension steps of the sequencing process which eliminates the errors introduced by molecular artifacts.
How does SBB sequencing work?
Next-level SBB sequencing chemistry proceeds by four primary steps: Initiation, Interrogation, Activation, and Incorporation.
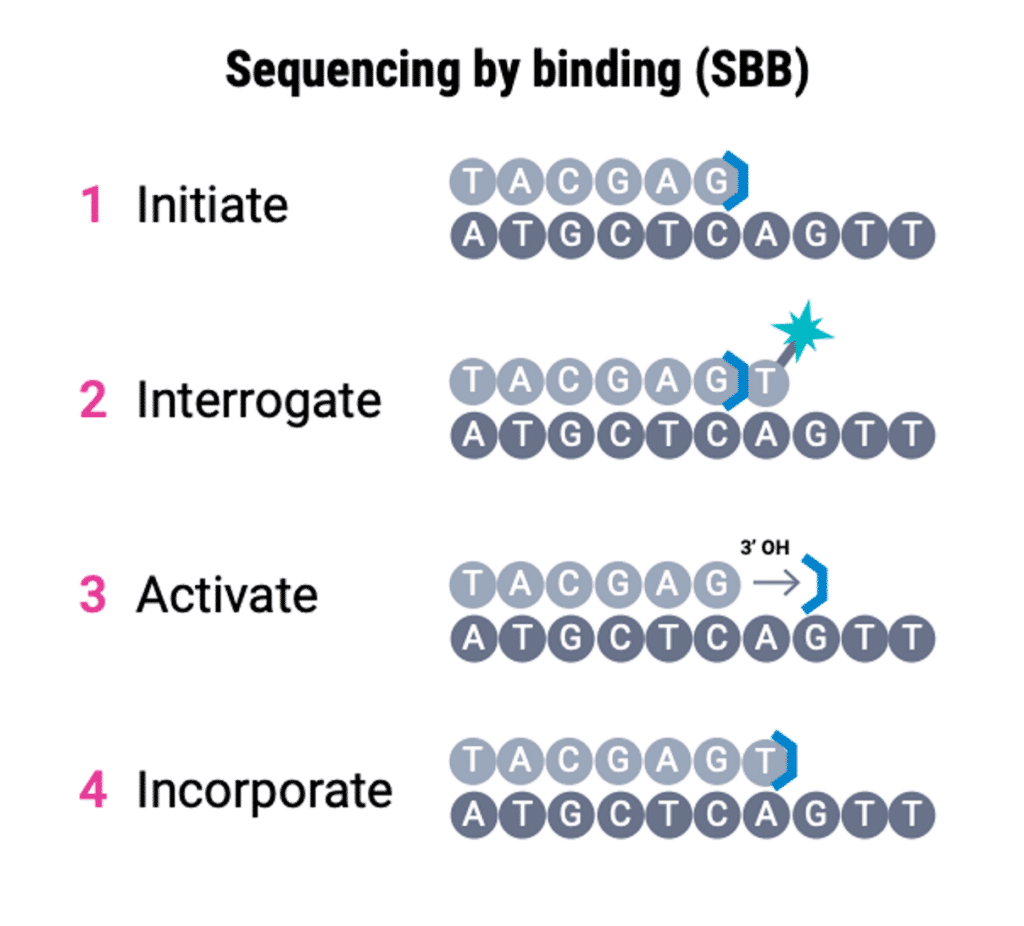
With SBB technology, sequencing is initiated by a polymerase on the DNA strand with a reversible blocker on the 3’ end to prevent additional bases from fully incorporating into this growing strand (step 1). Then, during interrogation fluorescently tagged bases flood the flow cell. Unlike traditional SBS sequencing, these tagged bases do not include a blocker. Once the appropriate tagged base binds to the DNA strand, a fluorescent signal is emitted, and this is measured using powerful optics (step 2). In a crucial difference from SBS sequencing, this tagged nucleotide base is not incorporated into the DNA strand but is washed away after the base signal is captured by the sequencing instrument. Thereafter, the 3’ end of the nucleotide is activated via the removal of the reversible blocker (step 3). This ensures the appropriate base will be incorporated from unlabeled, blocked nucleotides that flood the flow cell which blocks additional incorporation until the appropriate time in the next cycle (step 4).
How is SBB different from traditional SBS?
No molecular scarring
The key innovation behind the breakthrough accuracy of SBB sequencing lies in the fact that the fluorescently tagged nucleotide, which signals the base call, is not directly integrated into the DNA strand, as is typical in SBS sequencing. Separating these steps allows for separate optimization of the two steps, thereby enhancing the polymerase fidelity, signal-to-noise levels, and eliminating the detrimental effects of molecular scarring that occurs in conventional SBS sequencing when the fluorescent tag is removed from the growing DNA strand. With SBS sequencing, the cleaving needed to remove each fluorescent tag leaves residual linker arms attached to each nucleotide base (referred to as molecular scarring). Over the course of many cycles, these molecular residues cumulatively interfere with the polymerase as it attempts to read the DNA strand, which negatively affects the accuracy of the reads.
Minimal duplication
In addition to avoiding the accumulation of sequencing errors caused by molecular scarring, PacBio SBB sequencing has a much lower duplication rate than conventional short-read technology. Unlike reads that overlap, duplicated sequences add no additional information value to an analysis and are typically removed via bioinformatics in a process called deduplication. Because SBB sequencing creates fewer of these redundant sequences, less total data and computational resources are needed for the deduplication process, making for an overall more streamlined analysis.
Minimal index hopping
SBB sequencing causes minimal index hopping in multiplexed samples. Unlike SBB, the amplification methods of the patterned flow cells used by legacy SBS sequencing can cause a portion of unique indexes to cross over between sample pools during sequencing. This results in increased noise in the data caused by reads from one sample being erroneously assigned to the other. PacBio SBB sequencing does not have this problem.
Next-level accuracy
By eliminating molecular scarring, reducing duplication, and minimizing index hopping, SBB sequencing delivers extraordinary read accuracy with an unprecedented error rate of only 1 in 10,000 bases or less (Q40+).
What does Q40+ accuracy mean for the future of short-read sequencing?
With up to 15 × more accuracy than other bench top sequencers on the market†, SBB chemistry and the Onso system are revolutionizing what is possible with short-read sequencing technology.
But how is such game-changing accuracy measured and what can it enable you to do?
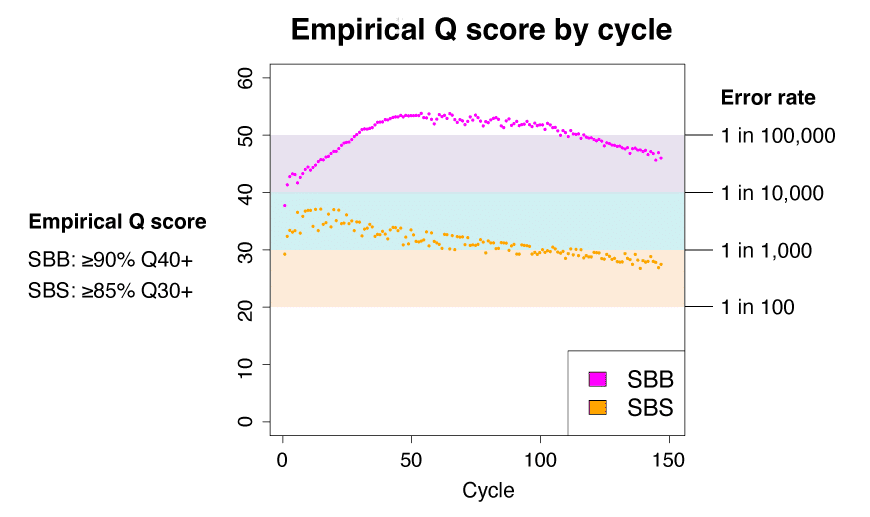
The accuracy of sequencing reads is measured using what genomics researchers call the Phred quality score, or Q score for short. Q scores are based on the likelihood of sequencing instrument making an incorrect base call. For example, a Q10 quality score represents a 1 in 10 error rate which translates to 90% accuracy. A score of Q20 represents a 1 in 100 error rate for 99% accuracy. Because this measure is represented in log scale, moving between these two quality scores represents a 10-fold jump in sequencing accuracy.
SBB sequencing on the PacBio Onso system returns 90% of bases at ≥Q40 which is a dramatic increase from the current Q30 scores reached by conventional SBS sequencing technologies. Remember: the transition from Q30 to Q40 accuracy means a jump from an error rate of 1 in 1,000 bases to only 1 in 10,000+! In many ways, an order of magnitude increase in accuracy holds the potential for an order of magnitude more discovery.
This remarkable Q40+ level of accuracy enables Onso users to more confidently sequence the genome and opens the door for great discoveries in research applications like liquid biopsy, gene editing, rare somatic variants, low-level resistance mutations in infectious disease, or anywhere else a needle in a haystack is sought to be found. The extraordinary sensitivity of SBB on the Onso system dramatically lowers the limit of detection for extremely rare-yet-critical variants missed by traditional short-read sequencing methods.
For example, the power of SBB sequencing enables researchers to perform up to 4-fold less sequencing than would be required using conventional SBS sequencing while maintaining the same or better sensitivity. SBB sequencing is also able to run samples onto flow cells at a 7 × lower concentration without impact on performance, enabling researchers to preserve more ever-so-valuable samples for future studies††.
Welcome to the era of accuracy
The amazing advantages of SBB chemistry represent a paradigm shift for the level of accuracy and sensitivity you can expect from a short-read sequencing technology. The value of Q40+ accuracy via SBB sequencing holds untold potential for discovery in applications of personal, population, and global health. With SBB, the era of accuracy has arrived.
Are you ready to try SBB + Onso?
Speak with a PacBio scientist
††Compared to NextSeq 1/2k and NovaSeq 6000 S1 input requirements with initial library pool concentration of 500-5000 pM. Assumes library insert size of 500 bp for mass calculation.