In the complex landscape of cancer biology, the exploration of microsatellite instability (MSI) has emerged as a promising frontier, offering potentially profound insights that could help improve diagnostic methodologies and therapeutic interventions. Microsatellites, a type of short tandem repeat (STR), are small repetitive DNA sequences scattered throughout the human genome that play a pivotal role in regulating gene expression and other cellular processes.1 However, when a cell’s DNA mismatch repair (MMR) system, a guardian of genomic integrity, falters, cells can lose their ability to regulate microsatellite lengths during division, resulting in MSI.2 Recent large-scale sequencing initiatives, particularly in cancer genomes, have begun to uncover the potential medical significance of MSI by revealing its prevalence in various tumor types, including colorectal, endometrial, and gastric adenocarcinomas.2
The importance of microsatellite instability in potentially guiding cancer treatment
In colorectal and endometrial cancers in particular, elevated levels of MSI in tumors have been shown to be a good predictor of a patient’s response to immunotherapy treatments.3,4 For this reason, research into the presence and implications of MSI across a diverse range of tumor types presents a prime opportunity for impactful advancements in cancer research. Clinical tests for MSI status in colorectal and endometrial cancers rely on either immunohistochemistry of MMR-related proteins or PCR of a small number of known microsatellite loci, but recent work has demonstrated the potential advantages of NGS (next generation sequencing) based approaches to help detect MSI in a much broader array of cancer types.5
However, the inherently repetitive nature of microsatellite sequences presents a significant challenge for conventional sequencing methods to correctly detect MSI. The use of NGS to help predict MSI status and potentially inform treatment strategies across cancer types thus relies on the sequencing instrument’s capacity to accurately read through these repetitive regions.
To tackle these challenges head-on, a team of scientists from the DOvEEgene (Detecting Ovarian and Endometrial cancer Early using Genomics) project at McGill University in Quebec, Canada recently explored the ability of the new PacBio Onso short-read sequencing system, with its extremely accurate (Q40+) sequencing by binding (SBB) chemistry, to try and find out whether it can more effectively detect MSI than standard approaches based on sequencing by synthesis (SBS). The preliminary results from this effort were presented at the Early Detection of Cancer conference in London in October 2023 and are highly encouraging.
DOvEEgene: Aiming to bring early detection capabilities to ovarian and endometrial cancer
The DOvEEgene project focuses on ovarian and endometrial cancers where a staggering 75% of cases are only identified in later, more advanced stages in the Canadian population. In response to the urgent need for early detection, the project was launched with the ambitious goal of identifying these cancers as early as possible. The team based at McGill University hopes to achieve this by developing a test that is not only low-cost and minimally invasive, but also widely accessible — much like how the Pap test revolutionized the detection of cervical cancer.
Highly accurate short-read sequencing as a new way forward in MSI detection strategies
The success of the DOvEEgene project hinges on the ability to accurately detect cancer variants with both very high sensitivity and specificity. To accomplish this, the researchers are leveraging the PacBio Onso system, which can achieve better performance in short tandem repeat regions compared to existing short-read sequencing technologies. Onso does this by decoupling the interrogation and incorporation steps of the sequencing cycle to reduce molecular scarring and improve polymerase performance, even in these challenging repetitive regions of a cancer genome.
More on Onso chemistry
To investigate the true capabilities of the Onso system against existing technology, DOvEEgene scientists collected intra-uterine brush samples alongside saliva samples from a group of test subjects. DNA was then extracted from these samples followed by library prep and hybrid capture using a panel targeting variants known to be involved in ovarian and endometrial cancers. Libraries were sequenced on both a NovaSeq (S4 flow cell) and the PacBio Onso sequencing platform. A machine learning algorithm, developed through extensive training and testing on a dataset of 481 samples, was then employed to predict the probability of a subject having cancer and to detect the associated MSI, based on the genomic data.
Gaining greater confidence in what you see and what you don’t
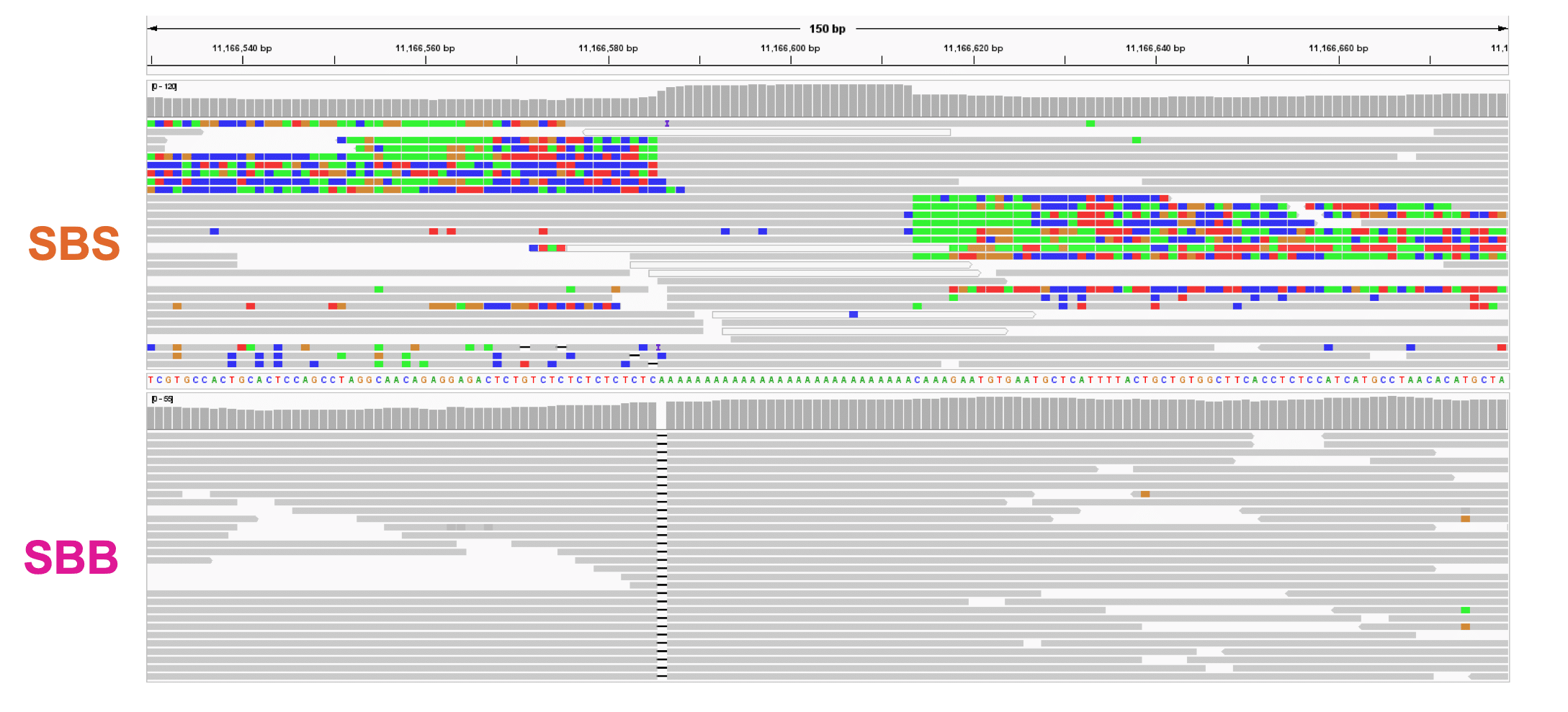
Although homopolymers, and poly-A repeats in particular, are the most frequent and mutation-prone type of microsatellite in the genome, existing sequencing methods for MSI detection have long struggled to accurately characterize these regions.6 As previously demonstrated on control samples, the Onso system’s powerful ability to sequence through homopolymers overcomes this limitation (an illustrative example from a separate comparative study performed on HG002 is shown in figure 1). It also reduces phasing issues leading to a more accurate enumeration of homopolymer length, which forms the basis for more accurately detecting MSI.
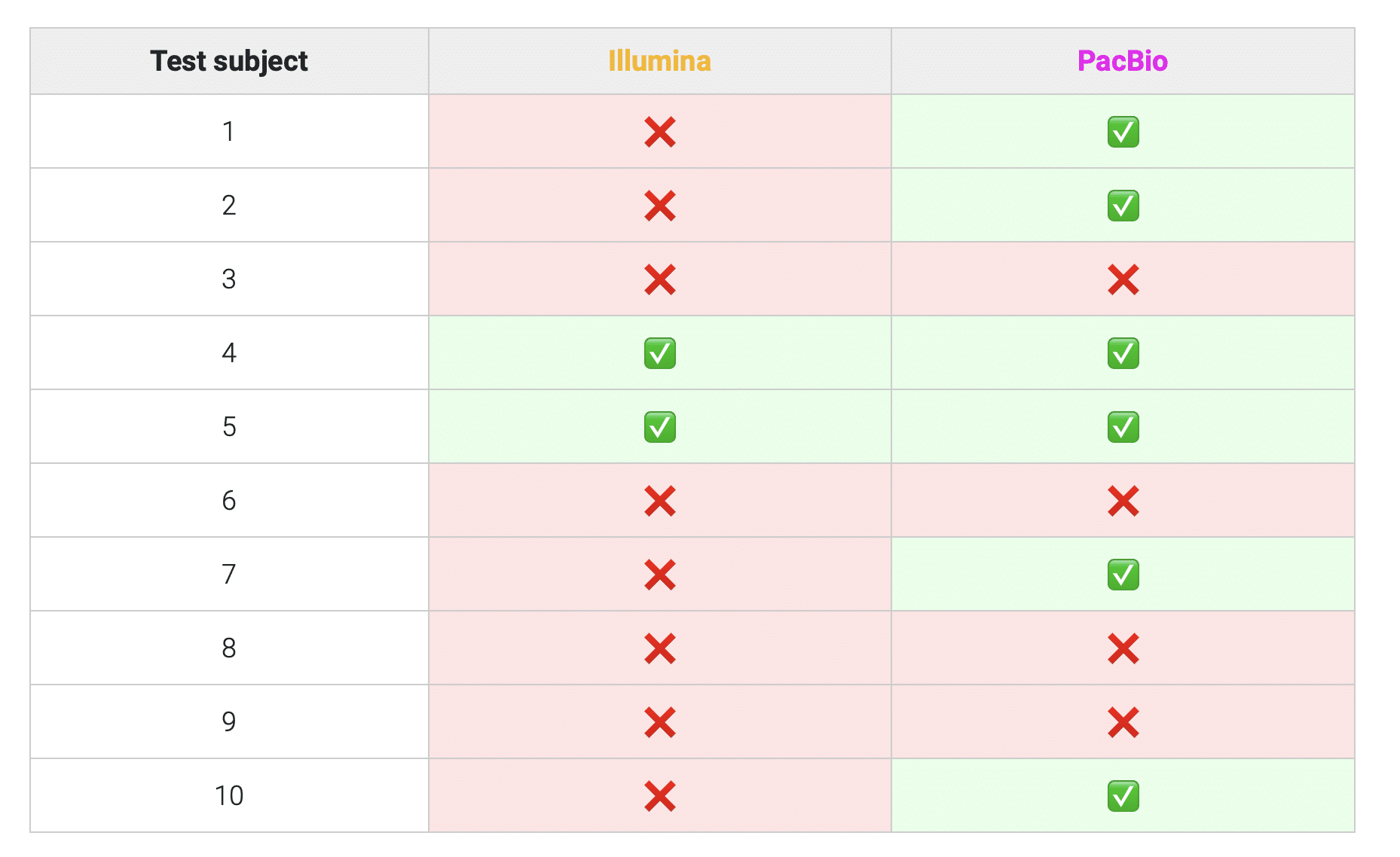
Preliminary data from the McGill team indicates that the PacBio Onso system identifies more unstable microsatellite loci in the DOvEEgene targeted panel regions than existing SBS methods, which could be a result of improved sequencing performance for Onso in these repetitive regions (table 1). While further work is needed to explore the basis for this increase in MSI detection with Onso, and to benchmark its performance against standard clinical tests, these findings from the McGill team suggest a promising future for Onso and its core SBB chemistry in research on early cancer detection, emphasizing the critical role that sequencing technologies play in improving the accuracy and reliability of applying genomic assays to a clinical setting in the future.
The continuous advancement of genomics and sequencing technologies, exemplified by the Onso system and SBB, have the potential to enable researchers to open new avenues of exploration in precision medicine and personalized cancer treatments. Stay tuned for the latest updates at AACR 2024 as we continue to support the cancer researcher community on its transformative path towards enabling earlier detection.
References
- Bagshaw ATM. Functional Mechanisms of Microsatellite DNA in Eukaryotic Genomes. Genome Biol Evol. 2017 Sep 1;9(9):2428-2443. doi: 10.1093/gbe/evx164. PMID: 28957459; PMCID: PMC5622345.
- Bonneville, R. et al. Landscape of Microsatellite Instability Across 39 Cancer Types. Precision Oncology. DOI: 10.1200/PO.17.00073. Published online October 3, 2017.
- Motta R, Cabezas-Camarero S, Torres-Mattos C, Riquelme A, Calle A, Figueroa A, Sotelo MJ. Immunotherapy in microsatellite instability metastatic colorectal cancer: Current status and future perspectives. J Clin Transl Res. 2021 Aug 4;7(4):511-522. PMID: 34541365; PMCID: PMC8445628.
- Green AK, Feinberg J, Makker V. A Review of Immune Checkpoint Blockade Therapy in Endometrial Cancer. Am Soc Clin Oncol Educ Book. 2020 Mar;40:1-7. doi: 10.1200/EDBK_280503. PMID: 32213091.
- Stephen J Salipante, Sheena M Scroggins, Heather L Hampel, Emily H Turner, Colin C Pritchard, Microsatellite Instability Detection by Next Generation Sequencing,Clinical Chemistry, Volume 60, Issue 9, 1 September 2014, Pages 1192–1199, https://doi.org/10.1373/clinchem.2014.223677
- Caitlin A. Loh, Danielle A. Shields, Adam Schwing, Gilad D. Evrony. High-fidelity, Large-scale Targeted Profiling of Microsatellites. bioRxiv 2023.11.28.569106; doi: https://doi.org/10.1101/2023.11.28.569106