 It’s not unusual for progeny to outperform their parents, and it’s often the goal in plant breeding. But tracing the molecular basis of such heterosis can be difficult, especially in diploid species with high genetic diversity and allele-specific expression like maize.
It’s not unusual for progeny to outperform their parents, and it’s often the goal in plant breeding. But tracing the molecular basis of such heterosis can be difficult, especially in diploid species with high genetic diversity and allele-specific expression like maize.
Cold Spring Harbor scientists have tackled the challenge using the PacBio Iso-Seq method and a new tool, IsoPhase.
As reported in Nature Communications Biology, Bo Wang, Doreen Ware, and colleagues performed an isoform-level phasing study in maize using the temperate line B73 and the tropical line Ki11, as well as their reciprocal crosses (B73 × Ki11; Ki11 × B73), which exhibit dramatic differences in height, root number and biomass from their parents.
The Cold Spring Harbor team phased 6,907 genes in the two reciprocal hybrids and were able to identify parental origin as well as novel isoforms in the hybrid lines. They also measured differing haplotypic expressions.
“Full-length, single-molecule sequencing provides an unprecedented allele-specific view of the haploid transcriptome,” the authors wrote.
“Haplotype phasing using long reads allowed us to accurately calculate allele-specific transcript and gene expression, as well as identify imprinted genes and investigate the cis/trans-regulatory effects.”
Because alleles from the same gene can generate heterozygous transcripts with distinct sequences, full analysis of allele-specific expression (ASE) is necessary to achieve a thorough understanding of transcriptome profiles. Previous attempts using short-read RNA-seq have provided expression information, but have not been able to provide full-length haplotype information.
The Cold Spring Harbor team used the Sequel platform to produce a single-molecule full-length cDNA dataset for the two maize parental lines and their reciprocal hybrid lines from root, embryo, and endosperm.
Barcoded SMRTbell libraries produced 4,898,979 HiFi reads, yielding 250,168 full-length, high-quality consensus transcript sequences. After mapping to the maize RefGen_v4 genome assembly and assessing for redundancy, the team ended up with 3,344 novel transcripts.
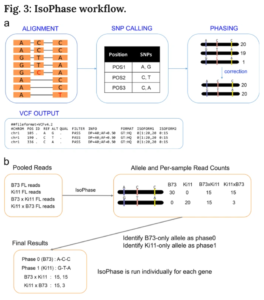
For phasing of these transcripts, the team applied the new IsoPhase tool, which uses the full-length nature of the reads and SNP calling to phase reads.
To determine which allele belonged to B73 or Ki11, they took advantage of the fact that all B73 reads must only express one allele, whereas all Ki11 reads must only express the other. Once the parental alleles were identified, they obtained the allelic counts for the F1 hybrids.
“Sequencing of full-length haplotype-specific isoforms enabled accurate assessment of allelic imbalance, which could be used to study the molecular mechanisms underlying genetic or epigenetic causative variants and associate expression polymorphisms with plant heterosis.
The approach does not require parental information (although parental data could be used to assign maternal and paternal alleles) and can be used on exclusively long-read data, they added.
“To our knowledge, this is the first full-length isoform phasing study in maize, or in any plant, and thus provides important information for haplotype phasing to other organisms, including polyploid species,” the authors wrote.
Learn more about PacBio transcriptome sequencing and the Iso-Seq analysis method.
July 17, 2020 | Agrigenomics