To enable better understanding of biology, sequencing data must be accurate and complete. This is especially true when seeking out variants and determining their implications.
Luckily, technical and software improvements for SMRT Sequencing are making it easier to efficiently generate genome assemblies with unparalleled accuracy.
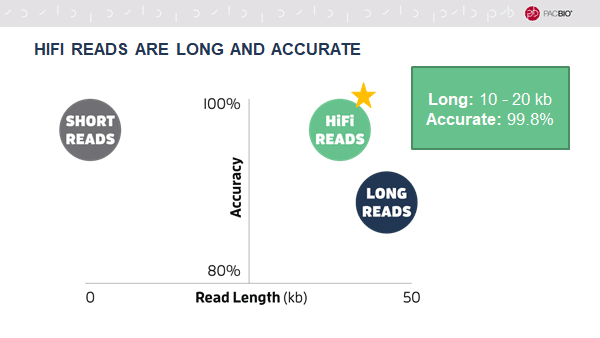
As presented in a webinar by PacBio Staff Scientist Sarah Kingan (@drsarahdoom) and GoogleAI Genomics Project Lead Andrew Carroll (@acarroll_ATG), HiFi reads enabled by circular consensus sequencing (CCS) on the new Sequel II System challenge the notion that sequencing technologies require a tradeoff between length and accuracy.
Kingan highlighted several benefits to using HiFi data for genome assembly:
- Higher accuracy of assemblies due to the high inherent base quality of HiFi reads
- Dramatic time-savings in generating a genome assembly
- Algorithmic improvement in the FALCON assembler that enhance the performance of HiFi assemblies

HiFi reads are extremely accurate because they utilize single-molecule consensus, rather than multiple-molecule consensus, which is required for traditional long-read assembly methods. The resulting HiFi assemblies have higher base accuracy than assemblies produced by continuous long reads.
HiFi reads are also more efficiently produced by CCS due to algorithmic enhancements that reduce compute time. CCS for a single SMRT Cell 8M run on the Sequel II System will be able to be completed in 3.5 hours with the upcoming software release.
Because the HiFi reads are already error corrected, the genome assembly process is simplified and streamlined, requiring only 20% of the compute time for a human genome compared to a continuous long read assembly.
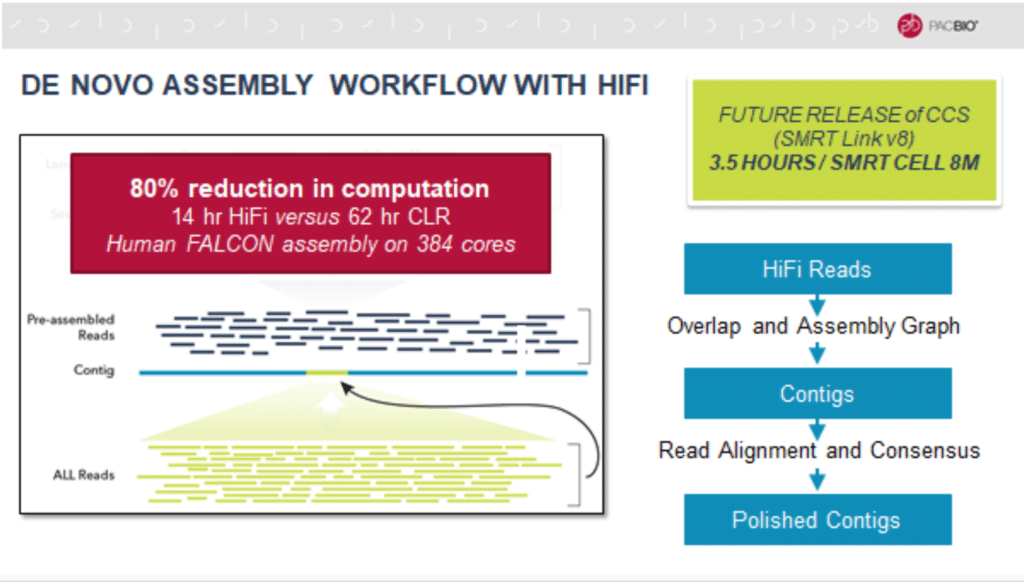
HiFi data needed HiFi-ready assembly tools
In order to make the most of these improvements, some assembly and analytical programs have also been modified.
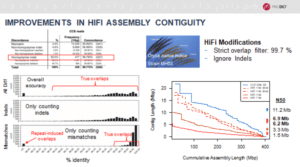
While testing the system on several human and animal genomes, Kingan said the PacBio team achieved equivalent or higher contiguity in multiple species, such as the fruit fly and bluefin tuna. But in a complex plant genome such as rice, with its multitude of repeat-induced overlaps, the results weren’t as robust.
So Kingan and colleagues modified the FALCON-Unzip assembler to make the most of the higher accuracy HiFi reads. By ignoring indel differences, they were able to better assemble the plant genomes. These latest features will be added soon to the already-incorporated improvements of faster read tracking and polishing.
Deep learning digs deeper
When it comes to assessing the “unknown unknowns,” artificial intelligence and machine learning is better than even the most robust human-designed algorithms, said GoogleAI’s Carroll.
His team has developed DeepVariant, a germline variant caller distinguished by its best-in-class accuracy. The open source program is also extensible – it can be re-trained for new technologies without writing new software — and this is exactly what his team did, in order to better handle HiFi data.
HiFi read errors are different from short-read errors, Carroll explained. Short-read data can lead to mapping complexity and coverage variability. HiFi reads are much more mappable and uniform, but can have noisier indel lengths in homopolymers, he said.
Carroll’s team fed the DeepVariant program millions of examples and labels from Genome in a Bottle to update weights in the model. Considering the range of uses and needs of PacBio users, they included data collected on both SMRT Cells 1M and 8M, featuring a variety of insert sizes and coverage levels.
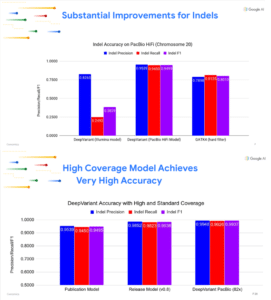
Better training yields better results
The team saw somewhat improved SNP accuracy, which was already very high; substantially improved indel accuracy; and robust, more uniform coverage titrations. The developers were surprised to see that DeepVariant was also able to call some structural variants without specifically being trained to do so. And the improved DeepVariant 8.0 was able to confidently call regions that were previously deemed “difficult,” “non-confident,” or “non-callable.”
“AI-based programs actually benefit from more data and more difficult – and different – data,” Carroll said. “There are thousands more variants we can now call confidently.” Improved haplotype phasing and the ability to call variants in other HiFi data types are also on the horizon, Carroll said.
Watch the complete webinar and visit www.pacb.com/HiFi to learn more: