
Population genomics initiatives are gaining traction around the world. There are currently more than 20 active programs spread around the globe, and these programs aim to sequence millions of genomes so that researchers can build diverse health databases, learn how biology and lifestyle affect health, and bring us closer to a future where precision medicine is the standard. While short-read sequencing technology has been the norm, a new preprint from research groups participating in the All of Us (AoU) program in the U.S. challenges the status quo, finding that HiFi long-read sequencing is, in fact, a better method to help move these initiatives forward.
Accuracy over read length for population genomics
Accuracy is a key component to the success of all population genomics programs. The databases being built will be relied upon for years to come and it is important to start with the best data possible. We recently posted about the overall significance of the preprint’s findings when it comes to the inherent value of HiFi technology. But, it doesn’t stop there.
The preprint shares the results of a technical pilot comparing short- and long-read sequencing. The pilot was conducted by researchers from groups at Baylor College of Medicine, the Broad Institute, Jackson Laboratory, Discovery Life Sciences, Harvard Medical School, Johns Hopkins University, Massachusetts General Hospital, the University of Washington, HudsonAlpha Institute for Biotechnology, and Rice University.
The finding? There are substantial differences in the ability to accurately sequence medically relevant genes with HiFi long reads producing the most accurate results for both small and large variants.
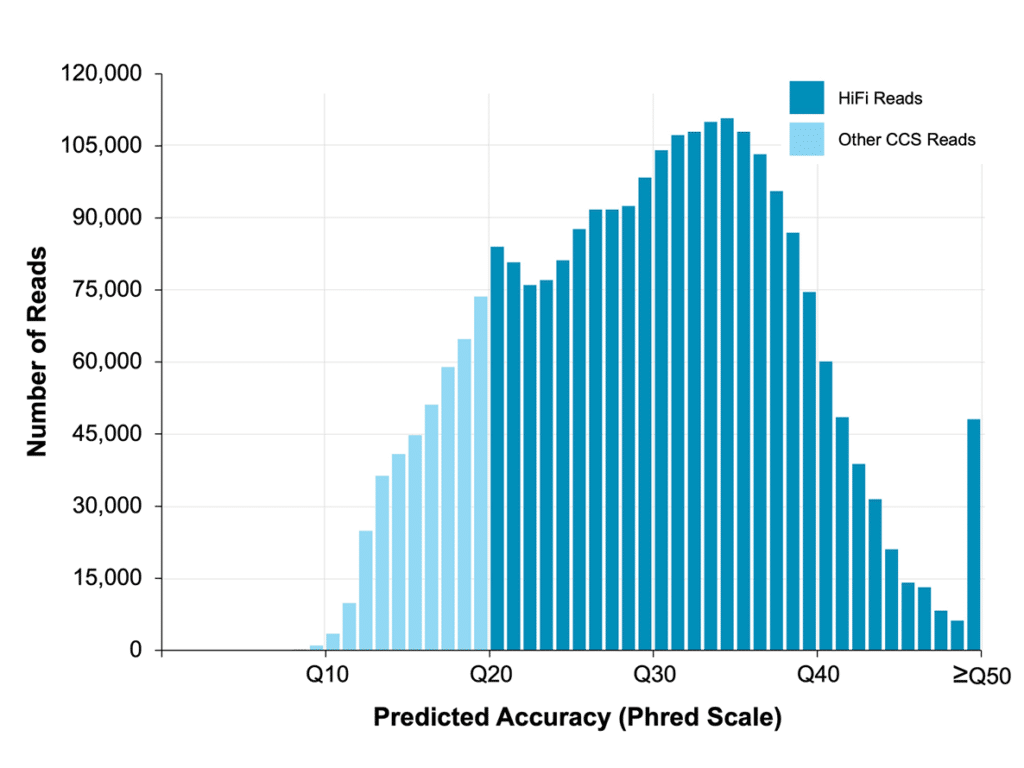
The pilot utilized a small cohort of samples from the HapMap project and two AoU control samples to investigate the utility of long-read sequencing for the AoU program. The authors noted that not only was HiFi sequencing more accurate, but single-read accuracy has a larger impact on variant calling ability than read length. And, in addition to the higher accuracy offered by HiFi reads, researchers can now also achieve lower costs and higher throughput thanks to technological advances with the new Revio system.
These findings have broad reach, and make it easy to see how the utility of HiFi sequencing can be significantly increased for population genomics studies, and thus precision medicine, to sequence medically relevant genes, find more variants at lower depth than legacy methods, and identify previously hidden variations.
HiFi long-read sequencing for missing heritability in genetic disorders
Another area where HiFi long-read sequencing is making a difference is through its ability to help close the gap on the issue of missing heritability in genetic disorders. Previously, variants detected in genome-wide association studies (GWAS) were not able to completely explain the heritability of complex traits. This is likely due to the inability to detect SVs (structural variants) properly.
SVs are much rarer (22-24,000 bp per genome) than single nucleotide variants (SNVs) (~4M bp or more per genome) but because of their larger sizes (>50 bp), SVs impact a larger number of base pairs per individual genome.
“SVs are an incredible source of genetic variation, but until recently remained inadequately understood because of the limits of sequencing technology,” said Fritz Sedlazeck, PhD, associate professor, Human Genome Sequencing Center at Baylor College of Medicine.Followed by,
“By utilizing long-read sequencing, we are now able to significantly improve the accuracy of SV detection and explain more biology.”The paper’s authors found the increased variant detection of HiFi sequencing enhanced SV calling and achieved a high accuracy of genome-wide SNV and indel calling. This in turn improves the power to link genetics to phenotypes of interests for novel discovery of genes and causative variants, allowing researchers to begin closing the gap on the missing heritability problem.
The authors conclude with,
“Longer term, the question rises if we have entered the age of using long reads exclusively. We conclude that despite currently scaling and costs considerations, we should continue developing population-scale cohorts sequenced with long reads only.”
These findings are just the first step in expanding the breadth and depth of data that can be created by population genomics programs around the world. If you would like to discuss how your program can benefit from HiFi long-read sequencing, please reach out to speak to a PacBio scientist.